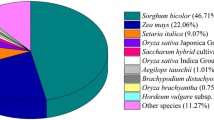
Abstract
Sugarcane (Saccharum officinarum L.) is the most important sugar crop and a major source of bio-ethanol. Sucrose accumulation is important in sugarcane, but it has not been investigated by transcriptome analysis . In this study, the transcriptome of a high-sucrose sugarcane variety, GT35, was sequenced using high-throughput Solexa technology. A total of 34,105,138 high-quality reads with an average length of 75 bp were obtained. These reads were assembled into 101,255 unigenes, with a mean length of 460 bp and an N50 length of 640 bp. BLAST searches indicated that 36,472 (36.0 %) unigenes had homologous sequences in the NCBI non-redundant protein sequences databases, and 23,811 (23.5 %) had homologous sequences in the Swiss-Prot databases. A total of 14,613 unigenes were assigned gene ontology terms, and 13,231 were assigned functional annotations and grouped into 25 functional categories. A KEGG pathway analysis of 30,756 unigenes revealed more than 30 pathways in the sugarcane transcriptome. A total of 3420 simple sequence repeats were identified in 3185 unigenes. The data in this study enriched the sugarcane transcriptome resources and will be useful for further comparison and functional genomic studies in genus Saccharum and family Poaceae, and will be useful for further detailed elucidation of sucrose accumulation in sugarcane.



Similar content being viewed by others
References
Cardoso-Silva, C.B., E.A. Costa, M.C. Mancini, T.W.A. Balsalobre, L.E.C. Canesin, L.R. Pinto, M.S. Carneiro, A.A.F. Garcia, A.P. Souza, and R. Vicentini. 2014. De novo assembly and transcriptome analysis of contrasting sugarcane varieties. PLoS One 9: e88462.
Chen, S., X. Huang, X. Yan, Y. Liang, Y. Wang, X. Li, X. Peng, X. Ma, L. Zhang, Y. Cai, T. Ma, L. Cheng, D. Qi, H. Zheng, X. Yang, X. Li, and G. Liu. 2013. Transcriptome analysis in sheepgrass (Leymus chinensis): A dominant perennial grass of the Eurasian steppe. PLoS One 8: e67974.
Chen, S., Y. Cai, L. Zhang, X. Yan, L. Cheng, D. Qi, Q. Zhou, X. Li, and G. Liu. 2014. Transcriptome analysis reveals common and distinct mechanisms for sheepgrass (Leymus chinensis) responses to defoliation compared to mechanical wounding. PLoS One 9: e89495.
Cheung, F., B.J. Haas, S.M. Goldberg, G.D. May, Y. Xiao, and C.D. Town. 2006. Sequencing Medicago truncatula expressed sequenced tags using 454 Life Sciences technology. BMC Genomics 7: 272.
Cho, Y.G., T. Ishii, S. Temnykh, X. Chen, L. Lipovich, S.R. McCouch, W.D. Park, N. Ayres, and S. Cartinhour. 2000. Diversity of microsatellites derived from genomic libraries and GenBank sequences in rice (Oryza sativa L.). Theoretical and Applied Genetics 100: 713–722.
Conesa, A., S. Götz, J.M. García-Gómez, J. Terol, M. Talón, and M. Robles. 2005. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21: 3674–3676.
Cordeiro, G.M., R. Casu, C.L. McIntyr, J.M. Manners, and R.J. Henry. 2001. Microsatellite markers from sugarcane (Saccharum spp.) ESTs cross transferable to erianthus and sorghum. Plant Science 160: 1115–1123.
D’Hont, A., D. Ison, K. Alix, C. Roux, and J.C. Glaszmann. 1998. Determination of basic chromosome numbers in the genus Saccharum by physical mapping of ribosomal RNA genes. Genome 41: 221–225.
D’Hont, A., L. Grivet, P.R.S. Feldmann, N. Berding, and J.C. Glaszmann. 1996. Characterisation of the double genome structure of modern sugarcane cultivars (Saccharum SPP.) by molecular cytogenetics. Molecular and General Genetics 250: 405–413.
da Maia, L.C., V.Q. de Souza, M.M. Kopp, F.I. de Carvalho, and A.C. de Oliveira. 2009. Tandem repeat distribution of gene transcripts in three plant families. Genetics and Molecular Biology 32: 822–833.
Deschamps, S., M.L. Rota, J.P. Ratashak, P. Biddle, D. Thureen, A. Farmer, S. Luck, M. Beatty, N. Nagasawa, L. Michael, V. Llaca, H. Sakai, G. May, J. Lightner, and M.A. Campbell. 2010. Rapid genome-wide single nucleotide polymorphism discovery in soybean and rice via deep resequencing of reduced representation libraries with the Illumina genome analyzer. Plant Genome 3: 53–68.
Dunckelman, P.H., and B.L. Legendre. 1982. Guide to sugarcane breeding in the template zone. USDA-ARS, ARM-S-22, New Orleans.
Emrich, S.J., W.B. Barbazuk, L. Li, and P.S. Schnable. 2007. Gene discovery and annotation using LCM-454 transcriptome sequencing. Genome Research 17: 69–73.
Feng, C., M. Chen, C.J. Xu, L. Bai, X.R. Yin, X. Li, A.C. Allan, I.B. Ferguson, and K.S. Chen. 2012. Transcriptomic analysis of Chinese bayberry (Myrica rubra) fruit development and ripening using RNA-seq. BMC Genomics 13: 19.
Garg, R., R.K. Patel, A.K. Tyagi, and M. Jain. 2011a. De novo assembly of chickpea transcriptome using short reads for gene discovery and marker identification. DNA Research 18: 53–63.
Garg, R., R.K. Patel, S. Jhanwar, P. Priya, A. Bhattacharjee, G. Yadav, S. Bhatia, D. Chattopadhyay, A.K. Tyagi, and M. Jain. 2011b. Gene discovery and tissue-specific transcriptome analysis in chickpea with massively parallel pyrosequencing and web resource development. Plant Physiology 156: 1661–1678.
Gene Ontology Consortium. 2008. The gene ontology project in 2008. Nucleic Acids Research 36: D440–D444.
Grabherr, M.G., B.J. Haas, M. Yassour, J.Z. Levin, D.A. Thompson, I. Amit, X. Adiconis, L. Fan, R. Raychowdhury, Q. Zeng, Z. Chen, E. Mauceli, N. Hacohen, A. Gnirke, N. Rhind, F. di Palma, B.W. Birren, C. Nusbaum, K. Lindblad-Toh, N. Friedman, and A. Regev. 2011. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nature Biotechnology 29: 644–652.
Gupta, P.K., S. Rustgi, S. Sharma, R. Singh, N. Kumar, and H.S. Balyan. 2003. Transferable EST-SSR markers for the study of polymorphism and genetic diversity in bread wheat. Molecular Genetics and Genomics 270: 315–323.
Hansey, C.N., B. Vaillancourt, R.S. Sekhon, N. de Leon, S.M. Kaeppler, and C.R. Buell. 2012. Maize (Zea mays L.) genome diversity as revealed by RNA-sequencing. PLoS One 7(3): e33071.
Hiremath, P.J., A. Farmer, S.B. Cannon, J. Woodward, H. Kudapa, R. Tuteja, A. Kumar, A. Bhanuprakash, B. Mulaosmanovic, N. Gujaria, L. Krishnamurthy, P.M. Gaur, P.B. Kavikishor, T. Shah, R. Srinivasan, M. Lohse, Y. Xiao, C.D. Town, D.R. Cook, G.D. May, and R.K. Varshney. 2011. Large-scale transcriptome analysis in chickpea (Cicer arietinum L.), an orphan legume crop of the semi-arid tropics of Asia and Africa. Plant Biotechnology Journal 9: 922–931.
Iseli, C., C.V. Jongeneel, and P. Bucher. 1999. ESTScan: A program for detecting, evaluating, and reconstructing potential coding regions in EST sequences. Proceedings of the International Conference on Intelligent Systems for Molecular Biology 7: 138–148.
Kanehisa, M., and S. Goto. 2000. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Research 28: 27–30.
Kanehisa, M., M. Araki, S. Goto, M. Hattori, M. Hirakawa, M. Itoh, T. Katayama, S. Kawashima, S. Okuda, T. Tokimatsu, and Y. Yamanishi. 2008. KEGG for linking genomes to life and the environment. Nucleic Acids Research 36: D480–D484 (Epub 2007 Dec 12).
Kanehisa, M., S. Goto, Y. Sato, M. Furumichi, and M. Tanabe. 2012. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Research 40: D109–D114.
La Rota, M., R.V. Kantety, J.K. Yu, and M.E. Sorrells. 2005. Nonrandom distribution and frequencies of genomic and EST-derived microsatellite markers in rice, wheat, and barley. BMC Genomics 6: 23.
Li, D., Z. Deng, B. Qin, X. Liu, and Z. Men. 2012. De novo assembly and characterization of bark transcriptome using Illumina sequencing and development of EST-SSR markers in rubber tree (Hevea brasiliensis Muell. Arg.). BMC Genomics 13: 192.
Li, Y.R. 2010. Modern sugarcane science. Beijing: China Agriculture Press.
Lister, R., R.C. O’Malley, J. Tonti-Filippini, B.D. Gregory, C.C. Berry, A.H. Millar, and J.R. Ecker. 2008. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133: 523–536.
Lu, T., G. Lu, D. Fan, C. Zhu, W. Li, Q. Zhao, Q. Feng, Y. Zhao, Y. Guo, W. Li, X. Huang, and B. Han. 2010. Function annotation of the rice transcriptome at single-nucleotide resolution by RNA-seq. Genome Research 20: 1238–1249.
Mao, X., T. Cai, J.G. Olyarchuk, and L. Wei. 2005. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 21: 3787–3793.
Mardis, E.R. 2008. The impact of next-generation sequencing technology on genetics. Trends in Genetics 24(3): 133–141.
Marguerat, S., and J. Bähler. 2010. RNA-seq: From technology to biology. Cellular and Molecular Life Sciences 67: 569–579.
Menossi, M., M.C. Silva-Filho, M. Vincentz, M.A. Van-Sluys, and G.M. Souza. 2008. Sugarcane functional genomics: gene discovery for agronomic trait development. International Journal of Plant Genomics 2008:458732. doi:10.1155/2008/458732
Moriya, Y., M. Itoh, S. Okuda, A.C. Yoshizawa, and M. Kanehisa. 2007. KAAS: An automatic genome annotation and pathway reconstruction server. Nucleic Acids Research 35: W182–W185.
Morozova, O., and M.A. Marra. 2008. Applications of next-generation sequencing technologies in functional genomics. Genomics 92: 255–264.
Ramu, P., B. Kassahun, S. Senthilvel, C. Ashok Kumar, B. Jayashree, R.T. Folkertsma, L.A. Reddy, M.S. Kuruvinashetti, B.I. Haussmann, and C.T. Hash. 2009. Exploiting rice-sorghum synteny for targeted development of EST-SSRs to enrich the sorghum genetic linkage map. Theoretical and Applied Genetics 119: 1193–1204.
Steuernagel, B., S. Taudien, H. Gundlach, M. Seidel, R. Ariyadasa, D. Schulte, A. Petzold, M. Felder, A. Graner, U. Scholz, K.F. Mayer, M. Platzer, and N. Stein. 2009. De novo 454 sequencing of barcoded BAC pools for comprehensive gene survey and genome analysis in the complex genome of barley. BMC Genomics 2009(10): 547.
Tatusov, R.L., M.Y. Galperin, D.A. Natale, and E.V. Koonin. 2000. The COG database: A tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Research 28: 33–36.
Thiel, T., W. Michalek, R.K. Varshney, and A. Graner. 2003. Exploiting EST databases for the development and characterization of gene-derived SSR markers in barley (Hordeum vulgare L.). Theoretical and Applied Genetics 106: 411–422.
Trick, M., Y. Long, J. Meng, and I. Bancroft. 2009. Single nucleotide polymorphism (SNP) discovery in the polyploid Brassica napus using Solexa transcriptome sequencing. Plant Biotechnology Journal 7: 334–346.
Troncoso-Ponce, M.A., A. Kilaru, X. Cao, T.P. Durrett, J. Fan, J.K. Jensen, N.A. Thrower, M. Pauly, C. Wilkerson, and J.B. Ohlrogge. 2011. Comparative deep transcriptional profiling of four developing oilseeds. Plant Journal 68: 1014–1027.
Varshney, R.K., S.N. Nayak, G.D. May, and S.A. Jackson. 2009. Next-generation sequencing technologies and their implications for crop genetics and breeding. Trends in Biotechnology 27: 522–530.
Victoria, F.C., L.C. da Maia, and A.C. de Oliveira. 2011. In silico comparative analysis of SSR markers in plants. BMC Plant Biology 11: 15.
Wang, W., Y. Wang, Q. Zhang, Y. Qi, and D. Guo. 2009. Global characterization of Artemisia annua glandular trichome transcriptome using 454 pyrosequencing. BMC Genomics 10: 465.
Weber, A.P.M., K.L. Weber, K. Carr, C. Wilkerson, and J.B. Ohlrogge. 2007. Sampling the Arabidopsis transcriptome with massively parallel pyrosequencing. Plant Physiology 144: 32–42.
Wei, W., X. Qi, L. Wang, Y. Zhang, W. Hua, D. Li, H. Lü, and X. Zhang. 2011. Characterization of the sesame (Sesamum indicum L.) global transcriptome using Illumina paired-end sequencing and development of EST-SSR markers. BMC Genomics 12: 451.
Wu, Q., L. Xu, J. Guo, Y. Su, and Y. Que. 2013. Transcriptome profile analysis of sugarcane responses to Sporisorium scitaminea infection using Solexa sequencing technology. Biomed Research International 2013: Article ID 298920.
Yang, S.S., Z.J. Tu, F. Cheung, W.W. Xu, J.F. Lamb, H.J. Jung, C.P. Vance, and J.W. Gronwald. 2011. Using RNA-seq for gene identification, polymorphism detection and transcript profiling in two alfalfa genotypes with divergent cell wall composition in stems. BMC Genomics 12: 199.
Ye, J., L. Fang, H. Zheng, Y. Zhang, J. Chen, Z. Zhang, J. Wang, S. Li, R. Li, L. Bolund, and J. Wang. 2006. WEGO: A web tool for plotting GO annotations. Nucleic Acids Research 34: W293–W297.
Zenoni, S., A. Ferrarini, E. Giacomelli, L. Xumerle, M. Fasoli, G. Malerba, D. Bellin, M. Pezzotti, and M. Delledonne. 2010. Characterization of transcriptional complexity during berry development in Vitis vinifera using RNA-seq. Plant Physiology 152: 1787–1795.
Zhang, J., C. Nagai, Q. Yu, Y.-B. Pan, T. Ayala-Silva, R.J. Schnell, J.C. Comstock, A.K. Arumuganathan, and R. Ming. 2012. Genome size variation in three Saccharum species. Euphytica 185: 511–519.
Acknowledgments
This work was supported by the National High-tech R&D Program (863 Program) (2013AA102604), the Natural Science Foundation of China (31160301, 31560415), the National Program for International Scientific Exchange (2013DFA31600), the Guangxi Special Funds for Bagui Scholars and Distinguished Scholars, the Natural Science Foundation of Guangxi (2011GXNSFF018002) and the Fundamental Research Fund of Guangxi Academy of Agriculture Sciences (2015YT03, 2014JZ02 and 2013YM41).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Dong-Liang Huang and Yi-Jing Gao have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Huang, DL., Gao, YJ., Gui, YY. et al. Transcriptome of High-Sucrose Sugarcane Variety GT35. Sugar Tech 18, 520–528 (2016). https://doi.org/10.1007/s12355-015-0420-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12355-015-0420-z