Abstract
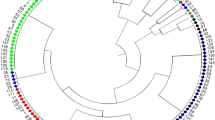
Molecular characterization and genetic diversity among 82 soybean accessions was carried out by using 44 simple sequence repeat (SSR) markers. Of the 44 SSR markers used, 40 markers were found polymorphic among 82 soybean accessions. These 40 polymorphic markers produced a total of 119 alleles, of which five were unique alleles and four alleles were rare. The allele number for each SSR locus varied between two to four with an average of 2.97 alleles per marker. Polymorphic information content values of SSRs ranged from 0.101 to 0.742 with an average of 0.477. Jaccard’s similarity coefficient was employed to study the molecular diversity of 82 soybean accessions. The pairwise genetic similarity among 82 soybean accessions varied from 0.28 to 0.90. The dendrogram constructed based on genetic similarities among 82 soybean accessions identified three major clusters. The majority of genotypes including four improved cultivars were grouped in a single subcluster IIIa of cluster III, indicating high genetic resemblance among soybean germplasm collection in India.


Similar content being viewed by others
References
Abe J, Xu DH, Suzuki Y, Kanazawa A, Shimamoto Y (2003) Soybean germplasm pools in Asia revealed by nuclear SSRs. Theor Appl Genet 106:445–453
Agarwal DK, Billore SD, Sharma AN, Dupare BU, Srivastava SK (2013) Soybean: introduction, improvement, and utilization in India—problems and prospects. Agric Res 2:293–300
Anonymous (2014) Annual report 2013–14, AICRP on Soybean, Directorate of Soybean Research, Indore, India.
Botstein D, White RL, Skolnick M, Davis RW (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphism. Am J Hum Genet 32:3
Cregan PB, Jarvik T, Bush AL, Shoemaker RC, Lark LG, Kahler AL (1999) An integrated linkage map of the soybean genome. Crop Sci 39:1464–1490
Diwan N, Cregan PB (1997) Automated sizing of fluorescent labeled simple sequence repeat (SSR) markers to assay genetic variation in soybean. Theor Appl Genet 95:723–733
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Du W, Wang M, Fu S, Yu D (2009) Mapping QTLs for seed yield and drought susceptiblity index in soybean (Glycine max L.) across different environments. J Genet Genom 36:721–731
Fu YB, Peterson GW, Morrison MJ (2007) Genetic diversity of Canadian soybean cultivars and exotic germplasm revealed by simple sequence repeat markers. Crop Sci 47:1947–1954
Guan R, Chang R, Li Y, Wang L, Liu Z, Qiu L (2010) Genetic diversity comparison between Chinese and Japanese soybeans (Glycine max (L.) Merr.) revealed by nuclear SSRs. Genet Resour Crop Evol 57:229–242
Hymowitz T (1970) On the domestication of the soybean. Econ Bot 24:408–421
Hymowitz T (1990) Soybeans: the success story. In: Janick J, Simon JE (eds) Advances in new crops: proceedings of the first national symposium on new crops: research, development, economics, held at Indianapolis, Ind., 23–26 October 1988. Timber Press, Portland, pp 159–163
Li W, Han Y, Zhang D, Yang M, Teng W, Jiang Z, Qiu L, Sun G (2008) Genetic diversity in soybean genotypes from north-eastern China and identification of candidate markers associated with maturity rating. Plant Breed 127:494–500
Liu et al (2011) QTL identification of flowering time at three different latitudes reveals homeologous genomic regions that control flowering in soybean. Theor Appl Genet 4:545–553
Maughan PJ, Saghai Maroof MA, Buss GR (1995) Microsatellite and amplified sequence length polymorphism in cultivated and wild soybean. Genome 38:715–725
Maughan PJ, Saghai Maroof MA, Buss GR, Huestis GM (1996) Amplified fragment length polymorphism (AFLP) in soybean: species diversity, inheritance and near isogenic line analysis. Theor Appl Genet 93:392–401
Narvel JM, Fehr WR, Chu WC, Grant D, Shoemaker RC (2000) Simple sequence repeat diversity among soybean plant introductions and elite genotypes. Crop Sci 40:1452–1458
Prabhakar, Bhatnagar PS (1995) Evaluation of soybean genetic resources. NRCS, Indore pp 1–96
Rohlf FJ (2000).. NTSYS-pc: Numerical taxonomy and multivariate analysis system, version 2.2. Exeter Software, Setauket, New York, USA.
Singh RK, Bhatia VS, Bhat KV, Mohapatra T, Singh NK, Bansal KC, Koundal KR (2010) SSR and AFLP based genetic diversity of soybean germplasm differing in photoperiod sensitivity. Genet and Mol Bio 33:319–324
Tantasawat P, Trongchuen J, Prajongjai T, Jenweerawat S, Chaowiset W (2011) SSR analysis of soybean (Glycine max (L.) Merr.) genetic relationship and variety identification in Thailand. Aust. J. Crop Sci 5:283–290
Thompson JA, Nelson RL (1998) Utilization of diverse germplasm for soybean yield improvement. Crop Sci 38:1362–1368
Tiwari SP, Joshi OP, Sharma AN (1999) The advent and renaissance of soybean: a landmark in Indian agriculture. NRCS, Indore, pp 1–58
Wang KJ, Takahata Y (2007) A preliminary comparative evaluation of genetic diversity between Chinese and Japanese wild soybean (Glycine soja) germplasm pools using SSR markers. Genet Resour Crop Evol 54:157–165
Wang L, Guan R, Zhangxiong L, Chang R, Qiu L (2006) Genetic diversity of Chinese cultivated soybean revealed by SSR markers. Crop Sci 46:1032–1038
Wang M, Li R, Yang W, Du W (2010) Assessing the genetic diversity of cultivars and wild soybeans using SSR markers. African J of Biotechnol 9:4857–4866
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Table 1
(DOCX 19 kb)
Rights and permissions
About this article
Cite this article
Kumawat, G., Singh, G., Gireesh, C. et al. Molecular characterization and genetic diversity analysis of soybean (Glycine max (L.) Merr.) germplasm accessions in India. Physiol Mol Biol Plants 21, 101–107 (2015). https://doi.org/10.1007/s12298-014-0266-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12298-014-0266-y