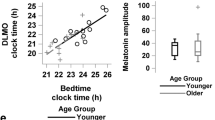
Abstract
The circadian rhythm of human circulating lipid components was studied under nearnormal tropical conditions in 162 healthy volunteers (103 males and 59 females; 7 to 75 years of age). They followed a diurnal activity from about 06:00 to about 22:00 and nocturnal rest. These volunteers were divided into four groups: Group A (7–20 years), Group B (21–40 years), Group C (41–60 years) and Group D (61–75 years), comprising 42, 60, 35 and 25 participants, respectively. A marked circadian rhythm was demonstrated for each studied variable in each group by population-mean cosinor analysis (almost invariably p < 0.001). Furthermore, circadian rhythm characteristics were compared among the 4 groups by parameter tests and regressed as a function of age, separately for males and females. A second-order polynomial characterized the MESOR of HDL cholesterol, phospholipids and total lipids, as well as the 24-h amplitude of total cholesterol and phospholipids. The 24-h amplitude of total lipids decreased linearly with age. The 24-h acrophase of the oldest age group (Group D) was advanced in the case of total cholesterol, HDL cholesterol, and total lipids, whereas that of phospholipids was delayed. Mapping the circadian rhythm (an important component of the broader time structure or chronome, which includes a. o., trends with age and extra-circadian components) of lipid components is needed to explore their role in the aging process in health.








Similar content being viewed by others
References
Liu JY, Li N, Yang J, Li N, Qiu H, Ai D, et al. Metabolic profiling of murine plasma reveals an unexpected biomarker in rofecoxib-mediated cardiovascular events. Proc Natl Acad Sci USA. 2010;107:17017–22.
Vouk K, Hevir N, Ribic´-Pucelj M, Haarpaintner G, Scherb H, Osredkar J, et al. Discovery of phosphatidylcholines and sphingomyelins as biomarkers for ovarian endometriosis. Hum Reprod. 2012;27:2955–65.
Sato Y, Suzuki I, Nakamura T, Bernier F, Aoshima K, Oda Y. Identification of a new plasma biomarker of Alzheimer’s disease using metabolomics technology. J Lipid Res. 2012;53:567–76.
Seijo S, Lozano JJ, Alonso C, Reverter E, Miquel R, Abraldes JG, et al. Metabolomics discloses potential biomarkers for the noninvasive diagnosis of idiopathic portal hypertension. Am J Gastroenterol. 2013;108:929–32.
Soga T, Baran R, Suematsu M, Ueno Y, Ikeda S, Sakarukawa T, et al. Differential metabolomics reveals ophthalmic acid as an oxidative stress biomarker indicating hepatic glutathione consumption. J Biol Chem. 2006;281:16768–76.
Beger RD, Sun J, Schnackenberg LK. Metabolomics approaches for discovering biomarkers of drug-induced hepatotoxicity and nephrotoxicity. Toxicol Appl Pharmacol. 2010;243:154–66.
Parman T, Bunin DI, Ng HH, McDunn JE, Wulff JE, Wang A, et al. Toxicogenomics and metabolomics of pentamethylchromanol (PMCol)-induced hepatotoxicity. Toxicol Sci. 2011;124:487–501.
McClay JL, Adkins DE, Vunck SA, Batman AM, Vann RE, Clark SL, et al. Large scale neurochemical metabolomics analysis identifies multiple compounds associated with methamphetamine exposure. Metabolomics. 2013;9:392–402.
Han X, Gross S. Shotgun lipidomics: electrospray ionization mass spectrometri analysis and quantitation of cellular lipidomes directly from crude extracts of biological samples. Mass Spectrom Rev. 2005;24:367–412.
Taguchi R, Nishijima M, Shimizu T. Basic analytical systems for lipidomics by mass spectrometry in Japan. Methods Enzymol. 2007;432:185–211.
Spiegel S, Milstien S. Sphingosine-1-phosphate: an enigmatic signaling lipid. Nat Rev Mol Cell Biol. 2003;4:397–407.
Serhan CN, Chiang N, Van Dyke TE. Resolving inflammation: dual antiinflammatoryand pro-resolution lipid mediators. Nat Rev Immunol. 2008;8:349–61.
Sparvero LJ, Amoscato AA, Kochanek PM, Pitt BR, Kagan VE, Bayir H. Mass-spectrometry based oxidative lipidomics and lipid imaging: applications in traumatic brain injury. J Neurochem. 2010;115:1322–36.
Han X, Rozen S, Boyle SH, Hellegers C, Cheng H, Burke JR, et al. Metabolomics in early Alzheimer’s disease: identification of altered plasma sphingolipidome using shotgun lipidomics. PLoS ONE. 2011;6:e21643.
Wang-Sattler R, Yu Z, Herder C, Messias AC, Floegel A, He Y, Heim K. Novel biomarkers for pre-diabetes identified by metabolomics. Mol Syst Biol. 2012;8:615.
Demirkan A, Isaacs A, Uqocsai P, Liebisch G, Struchalin M, Rudan I, et al. Plasmaphosphatidylcholine and sphingomyelin concentrations are associated withdepression and anxiety symptoms in a Dutch family-based lipidomics study. J Psychiatr Res. 2013;47:357–62.
Wishart DS, Jewison T, Guo AC, Wilson M, Knox C, Liu Y, et al. HMDB 3.0—the human metabolome database in 2013. Nucleic Acid Res. 2013;41:D801–7.
Rivera-Coll A, Fuentes-Arderiu X, Díez-Noguera A. Circadian rhythmic variations in serum concentrations of clinically important lipids. Clin Chem. 1994;40:1549–53.
Kessler Cella L, Van Cauter E, Schoeller DA. Diurnal rhythmicity of human cholesterol synthesis: normal pattern and adaptation to simulated “jet lag”. Am J Physiol. 1995;269:E489–98.
Kessler Cella L, Van Cauter E, Schoeller DA. Effect of meal timing on diurnal rhythm of human cholesterol synthesis. Am J Physiol. 1995;269:E878–80.
Singh RK, Wu J, Zhou S, Halberg F. Circadian rhythmic human circulating cholesterol in health, during fasting and on vegetarian vs. omnivorous diets. Chronobiologia. 1989;16:183.
Singh RK, Mahdi AA, Singh AK, Bansal SK, Wu J, Zhou S, Halberg F. Circadian variation of human circulating cholesterol components on vegetarian and omnivorous diets in healthy Indians. Indian J Clin Biochem. 1992;7:185–92.
Cornelissen G, Galli C, Halberg F, Meester FDE, Rise P, Wilczynska-Kwiatek A, et al. Circadian time structure of fatty acids and vascular monitoring. J Applied Biomed. 2010;8:93–109.
Halberg F. Chronobiology: methodological problems. Acta Med Rom. 1980;18:399–440.
Cornelissen G, Halberg F. Chronomedicine. In: Armitage P, Colton T, editors. Encyclopedia of biostatistics. 2nd ed. Chichester: Wiley; 2005. pp. 796–812.
Refinetti R, Cornelissen G, Halberg F. Procedures for numerical analysis of circadian rhythms. Biological Rhythm Res. 2007;38(4):275–325.
Cornelissen G. Cosinor-based rhythmometry. Theoretical Biology and Medical Modelling. 2014;11:16–40. doi:10.1186/1742-4682-11-16.24.
Bingham C, Arbogast B, Cornelissen G, Lee JK, Halberg F. Inferential statistical methods for estimating and comparing cosinor parameters. Chronobiologia. 1982;9:397–439.
Isabelle C, Josaiane S, Thomas M, Gerard S. Variations in total phospholipid and HDL-phospholipids in plasma from a general population: reference intervals and influence of xenobiotics. Clin Chem. 1985;31(5):763–6.
Kochhar S, Doris M. Jacobs, Ziad Ramadan. Probing gender-specific metabolism differences in humans by nuclear magnetic resonance-based metabonomics. Anal Biochem. 2006;352(2):274–81.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
None.
Rights and permissions
About this article
Cite this article
Singh, R., Sharma, S., Singh, R.K. et al. Circadian Time Structure of Circulating Plasma Lipid Components in Healthy Indians of Different Age Groups. Ind J Clin Biochem 31, 215–223 (2016). https://doi.org/10.1007/s12291-015-0519-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12291-015-0519-8