Abstract
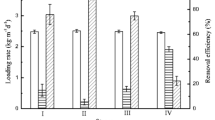
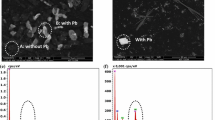
Sulfate-reducing bacteria (SRB) are widely used for heavy metal (HM) treatment in bioreactors but their growth and biological activity can be inhibited by such treatment. Here, bioreactor experiments were used to investigate changes in the SRB community and the copy number of the dissimilatory sulfite reductase β-subunit functional gene (dsrB) under high doses of sulfates and HMs. The SRB community was investigated using polymerase chain reaction denaturing gradient gel electrophoresis (PCR-DGGE) and sequencing techniques, while the dsrB gene abundance was measured by quantitative real-time PCR (qRT-PCR). The sulfate reduction rate was initially much higher in reactors without HMs than in those containing HMs (p = 0.001). Sulfate levels were reduced by 50% within the first 3 days of operation. As a result, the HM removal rate was initially much lower in the reactors containing HMs. Most of the HMs reduced to safe limits within 9 ~ 12 days of operation. The SRB community mainly consisted of Desulfovibrio vulgaris, D. termitidis, D. desulfuricans, D. simplex and Desulfomicrobium baculatum, as determined by PCR-DGGE. qRT-PCR revealed a decreasing trend in the copy numbers of a functional gene (dsrB) after 6 days in samples lacking HMs; however, the opposite trend was observed in the HM-containing samples.
Similar content being viewed by others
References
Berghorn, G. H. and G. R. Hunzeker (2001) Passive treatment alternatives for remediating abandoned-mine drainage. Remed. J. 11: 111–127.
Waybrant, K. R., C. J. Ptacek, and D. W. Blowes (2002) Treatment of mine drainage using permeable reactive barriers: Column experiments. Environ. Sci. Technol. 36: 1349–1356.
Hiibel, S. R., L. P. Pereyra, L. Y. Inman, A. Tischer, D. J. Reisman, K. F. Reardon, and A. Pruden (2008) Microbial community analysis of two field-scale sulfate-reducing bioreactors treating mine drainage. Environ. Microbiol. 10: 2087–2097.
Johnson, D. B. and K. B. Hallberg (2005) Acid mine drainage remediation options: A review. Sci. Total Environ. 338: 3–14.
Fang, H. H. P., L.-C. Xu, and K.-Y. Chan (2002) Effects of toxic metals and chemicals on biofilm and biocorrosion. Water Res. 36: 4709–4716.
Hao, O. J., L. Huang, J. M. Chen, and R. L. Buglass (1994) Effects of metal additions on sulfate reduction activity in wastewaters. Toxicol. Environ. Chem. 46: 197–212.
Poulson, S. R., P. J. S. Colberg and J. I. Drever (1997) Toxicity of heavy metals (Ni, Zn) to desulfovibrio desulfuricans. Geomicrobiol. J. 14: 41–49.
Alvarez, M. T., C. Crespo, and B. Mattiasson (2007) Precipitation of Zn (II), Cu (II) and Pb (II) at bench-scale using biogenic hydrogen sulfide from the utilization of volatile fatty acids. Chemosphere. 66: 1677–1683.
Jiménez-Rodríguez, A. M., M. M. Durán-Barrantes, R. Borja, E. Sánchez, M. F. Colmenarejo, and F. Raposo (2009) Heavy metals removal from acid mine drainage water using biogenic hydrogen sulphide and effluent from anaerobic treatment: Effect of pH. J. Hazard. Mater. 165: 759–765.
Çetin, D., S. Dönmez, and G. Dönmez (2008) The treatment of textile wastewater including chromium(VI) and reactive dye by sulfate-reducing bacterial enrichment. J. Environ. Manage. 88: 76–82.
Tebo, B. M. and A. Y. Obraztsova (1998) Sulfate-reducing bacterium grows with Cr(VI), U(VI), Mn(IV), and Fe(III) as electron acceptors. FEMS Microbiol. Lett. 162: 193–198.
Geets, J., K. Vanbroekhoven, B. Borremans, J. Vangronsveld, L. Diels, and D. van der Lelie (2006) Column experiments to assess the effects of electron donors on the efficiency of in situ precipitation of Zn, Cd, Co and Ni in contaminated groundwater applying the biological sulfate removal technology. Environ. Sci. Pollut. Res. 13: 362–378.
Rogers, S. and N. McClure (2003) The role of microbiological studies in bioremediation process optimization. pp. 27–59. Im, H. S. I. and M. Mg (eds.)., Bioremediation: A critical review. Horizon Scientific Press, Wymondham, UK.
Mackie, R. I., P. G. Stroot, and V. H. Varel (1998) Biochemical identification and biological origin of key odor components in livestock waste. J. Anim. Sci. 76: 1331–1342.
Spence, C., T. R. Whitehead, and M. A. Cotta (2008) Development and comparison of SYBR Green quantitative real-time PCR assays for detection and enumeration of sulfate-reducing bacteria in stored swine manure. J. Appl. Microbiol. 105: 2143–2152.
Kjeldsen, K. U., A. Loy, T. F. Jakobsen, T. R. Thomsen, M. Wagner, and K. Ingvorsen (2007) Diversity of sulfate-reducing bacteria from an extreme hypersaline sediment, Great Salt Lake (Utah). FEMS Microbiol. Ecol. 60: 287–298.
Kondo, R., D. B. Nedwell, K. J. Purdy, and S. Q. Silva (2004) Detection and enumeration of sulphate-reducing bacteria in estuarine sediments by competitive PCR. Geomicrobiol. J. 21: 145–157.
Leloup, J., A. Loy, N. J. Knab, C. Borowski, M. Wagner, and B. B. Jorgensen (2007) Diversity and abundance of sulfate-reducing microorganisms in the sulfate and methane zones of a marine sediment, Black Sea. Environ. Microbiol. 9: 131–142.
Pérez-Jiménez, J. R., L. Y. Young, and L. J. Kerkhof (2001) Molecular characterization of sulfate-reducing bacteria in anaerobic hydrocarbon-degrading consortia and pure cultures using the dissimilatory sulfite reductase (dsrAB) genes. FEMS Microbiol. Ecol. 35: 145–150.
Foti, M., D. Y. Sorokin, B. Lomans, M. Mussman, E. E. Zacharova, N. V. Pimenov, J. G. Kuenen, and G. Muyzer (2007) Diversity, activity, and abundance of sulfate-reducing bacteria in saline and hypersaline soda lakes. Appl. Environ. Microbiol. 73: 2093–2100.
Minz, D., J. L. Flax, S. J. Green, G. Muyzer, Y. Cohen, M. Wagner, B. E. Rittmann, and D. A. Stahl (1999) Diversity of sulfatereducing bacteria in oxic and anoxic regions of a microbial mat characterized by comparative analysis of dissimilatory sulfite reductase genes. Appl. Environ. Microbiol. 65: 4666–4671.
Dhillon, A., A. Teske, J. Dillon, D. A. Stahl, and M. L. Sogin (2003) Molecular characterization of sulfate-reducing bacteria in the guaymas basin. Appl. Environ. Microbiol. 69: 2765–2772.
Wagner, M., A. J. Roger, J. L. Flax, G. A. Brusseau, and D. A. Stahl (1998) Phylogeny of dissimilatory sulfite reductases supports an early origin of sulfate respiration. J. Bacteriol. 180: 2975–2982.
Atlas, R. M. (2005) Handbook of Media for Environmental Microbiology. CRC Press, Taylor & Francis Group., Boca Raton, FL, USA.
APHA, AWWA and WEF (1998) Standard Methods for the Examination of Water and Wastewater. p.1325. 20th ed. American Public Health Association, Washington D. C., USA.
Dar, S. A., L. Yao, U. van Dongen, J. G. Kuenen, and G. Muyzer (2007) Analysis of diversity and activity of sulfate-reducing bacterial communities in sulfidogenic bioreactors using 16S rRNA and dsrB genes as molecular markers. Appl. Environ. Microbiol. 73: 594–604.
Chervoneva, I., T. Hyslop, B. Iglewicz, L. Johns, H. R. Wolfe, S. Schulz, E. Leong, and S. Waldman (2006) Statistical algorithm for assuring similar efficiency in standards and samples for absolute quantification by real-time reverse transcription polymerase chain reaction. Anal. Biochem. 348: 198–208.
Perry, R. H. and D. W. Green (2008) Perry’s Chemical Engineers’ Handbook. 8th Edition, McGraw-Hill.
O’Flaherty, V. and E. Colleran (1999) Effect of sulphate addition on volatile fatty acid and ethanol degradation in an anaerobic hybrid reactor. I: Process disturbance and remediation. Bioresour. Technol. 68: 101–107.
MEP (2002) Environmental quality standard for surface water. GB 3838-2002, Ministry of Environmental Protection, P. R. China.
Shuttleworth, K. L. and R. F. Unz (1991) Influence of metals and metal speciation on the growth of filamentous bacteria. Water Res. 25: 1177–1186.
Al-Zuhair, S., M. H. El-Naas and H. Al-Hassani (2008) Sulfate inhibition effect on sulfate reducing bacteria. J. Biochem. Technol. 1: 39–44.
Kirchman, D. L. and J. H. Rich (1997) Regulation of bacterial growth rates by dissolved organic carbon and temperature in the equatorial pacific ocean. Microb. Ecol. 33: 11–20.
Rzeczycka, M. and M. Blaszczyk (2005) Growth and activity of aulphate-reducing bacteria in media containing phosphogypsum and different sources of carbon. Pol. J. Environ. Stud. 14: 891–895.
Zhang, W., L. S. Song, J. S. Ki, C. K. Lau, X. D. Li, and P. Y. Qian (2008) Microbial diversity in polluted harbor sediments II: Sulfate-reducing bacterial community assessment using terminal restriction fragment length polymorphism and clone library of dsrAB gene. Estuar. Coast Shelf Sci. 76: 682–691.
Cabrera, G., R. Perez, J. M. Gomez, A. Abalos, and D. Cantero (2006) Toxic effects of dissolved heavy metals on Desulfovibrio vulgaris and Desulfovibrio sp. strains. J. Hazard. Mater. 135: 40–46.
Quillet, L., L. Besaury, M. Popova, S. Paisse, J. Deloffre, and B. Ouddane (2012) Abundance, diversity and activity of sulfatereducing prokaryotes in heavy metalcontaminated sediment from a salt marsh in the Medway Estuary (UK). Mar. Biotechnol. 14: 363–381.
Lloyd, J. R., A. N. Mabbett, D. R. Williams, and L. E. Macaskie (2001) Metal reduction by sulphate-reducing bacteria: Physiological diversity and metal specificity Hydrometal. 59: 327–337.
Andrade, L. L., D. Leite, E. Ferreira, L. Ferreira, G. R. Paula, M. Maguire, and C. R. Hubert (2012) Microbial diversity and anaerobic hydrocarbon degradation potential in an oil-contaminated mangrove sediment. BMC Microbiol. 12: 186.
Varon-Lopez, M., A. C. F. Dias, C. C. Fasanella, A. Durrer, I. S. Melo, E. E. Kuramae, and F. D. Andreote (2013) Sulphur-oxidizing and sulphate-reducing communities in Brazilian mangrove sediments. Environ. Microbiol. doi:10.1111/1462-2920.12237.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Islamud-Din, Hesham, A.EL., Ahmad, A. et al. PCR-DGGE and real-time PCR dsrB-based study of the impact of heavy metals on the diversity and abundance of sulfate-reducing bacteria. Biotechnol Bioproc E 19, 703–710 (2014). https://doi.org/10.1007/s12257-014-0324-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12257-014-0324-x