Abstract
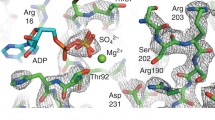
Microtubule not only provides the track for kinesin but also modulates kinesin’s mechanochemical cycle. Microtubule binding greatly increases the rates of two chemical steps occurring inside the nucleotide-binding pocket (NBP) of kinesin, i.e., ATP hydrolysis and ADP release. Kinesin neck linker docking (the key force-generation step) is initiated by the motor head rotation induced by ATP binding which needs an anchor provided by microtubule. These functions of microtubule can only be accomplished through interactions with kinesin. Based on the newly obtained crystal structures of kinesin-microtubule complexes, we investigate the interactions between kinesin’s NBP and microtubule using molecular dynamics simulations. We find that the N-3 motif of NBP has direct interactions with a group of negatively charged residues on α-tubulin through Ser235 and Lys237. These specific long-range interactions induce binding of NBP to microtubule at the right position and assist the formation of the indirect interaction between NBP and microtubule. These interactions between N-3 and microtubule have an important anchor effect for kinesin’s motor domain during its rotation with Ser235 as the rotation center, and also play a crucial role in stabilizing the ATP-hydrolysis environment.






Similar content being viewed by others
References
Asenjo, A. B., N. Krohn and H. Sosa. Configuration of the two kinesin motor domains during ATP hydrolysis. Nat. Struct. Mol. Biol. 10:836–842, 2003.
Asenjo, A. B. and H. Sosa. A mobile kinesin-head intermediate during the ATP-waiting state. Proc. Natl. Acad. Sci. USA 106:5657–5662, 2009.
Asenjo, A. B., Y. Weinberg and H. Sosa. Nucleotide binding and hydrolysis induces a disorder-order transition in the kinesin neck-linker region. Nat. Struct. Mol. Biol. 13:648–654, 2006.
Atherton, J., I. Farabella, I.-M. Yu, S. S. Rosenfeld, A. Houdusse, et al.. Conserved mechanisms of microtubule-stimulated ADP release, ATP binding, and force generation in transport kinesins. eLife 3:e03680, 2014.
Block, S. M. Kinesin motor mechanics: Binding, stepping, tracking, gating, and limping. Biophys. J. 92:2986–2995, 2007.
Cao, L., W. Wang, Q. Jiang, C. Wang, M. Knossow, et al. The structure of apo-kinesin bound to tubulin links the nucleotide cycle to movement. Nat. Commun. 5:5364, 2014.
Crevel, I., A. Lockhart and R. A. Cross. Weak and strong states of kinesin and ncd. J. Mol. Biol. 257:66–76, 1996.
Geng, Y., Q. Ji, S. Liu and S. Yan. Initial conformation of kinesin’s neck linker. Chin. Phys. B 23:108701, 2014.
Geng, Y., S. Liu, Q. Ji and S. Yan. Mechanical amplification mechanism of kinesin’s β-domain. Arch Biochem. Biophys. 543:10–14, 2014.
Gigant, B., W. Wang, B. Dreier, Q. Jiang, L. Pecqueur, et al. Structure of a kinesin-tubulin complex and implications for kinesin motility. Nat. Struct. Mol. Biol. 20:1001–1007, 2013.
Hackney, D. D. Kinesin atpase: rate-limiting ADP release. Proc Natl Acad Sci USA 85: 6314–6318, 1988.
Hancock, W. O. and J. Howard. Processivity of the motor protein kinesin requires two heads. J. Cell Biol. 140:1395–1405, 1998.
Hirokawa N., S. Niwa and Y. Tanaka. Molecular motors in neurons: Transport mechanisms and roles in brain function, development, and disease. Neuron 68:610–638, 2010.
Hirokawa N., and Y. Noda. Intracellular transport and kinesin superfamily proteins, kifs: Structure, function, and dynamics. Physiol. Rev. 88:1089–1118, 2008.
Howard, J. Mechanics of Motor Proteins and the Cytoskeleton. Sunderland: Sinauer Associates Inc, 2001.
Humphrey W., A. Dalke, and K. Schulten. Vmd: Visual molecular dynamics. J. Mol. Graphics 14:33–38, 1996.
Jorgensen W.L., J. Chandrasekhar, J.D. Madura, R.W. Impey, and M.L. Klein. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79:926–935, 1983.
Kanada, R., T. Kuwata, H. Kenzaki, S. Takada. Structure-based molecular simulations reveal the enhancement of biased brownian motions in single-headed kinesin. PLoS Comput. Biol. 9:e10022907, 2013.
Kikkawa, M., and N. Hirokawa. High-resolution cryo-EM maps show the nucleotide binding pocket of KIF1A in open and closed conformations. EMBO J. 25:4187–4194, 2006.
Kikkawa M., E. P. Sablin, Y. Okada, H. Yajima, R. J. Fletterick, et al. Switch-based mechanism of kinesin motors. Nature 411:439–445, 2001.
Krukau, A., V. Knecht and R. Lipowsky. Allosteric control of kinesin’s motor domain by tubulin: a molecular dynamics study. Phys. Chem. Chem. Phys. 16:6189–6198, 2014.
Kull, F. J., E. P. Sablin, R. Lau, R. J. Fletterick and R. D. Vale. Crystal structure of the kinesin motor domain reveals a structural similarity to myosin. Nature 380:550–555, 1996.
Lawrence C. J., R. K. Dawe, K. R. Christie, D. W. Cleveland, and S. C. Dawson, et al. A standardized kinesin nomenclature. J. Cell Biol. 167:19–22, 2004.
Li, M. and W. Zheng. All-atom structural investigation of kinesin-microtubule complex constrained by high-quality cryo-electron-microscopy maps. Biochemistry 51:5022–5032, 2012.
MacKerell A. D., D. Bashford, Bellott, R. L. Dunbrack, J. D. Evanseck, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 102:3586–3616, 1998.
McGrath, M. J., I.-F. W. Kuo, S. Hayashi and S. Takada. Adenosine triphosphate hydrolysis mechanism in kinesin studied by combined quantum-mechanical/molecular-mechanical metadynamics simulations. J. Am. Chem. Soc. 135:8908–8919, 2013.
Naber, N., T. J. Minehardt, S. Rice, X. Chen, J. Grammer, et al. Closing of the nucleotide pocket of kinesin-family motors upon binding to microtubules. Science 300:798–801, 2003.
Naber, N., S. Rice, M. Matuska, R. D. Vale, R. Cooke, et al. EPR spectroscopy shows a microtubule-dependent conformational change in the kinesin switch 1 domain. Biophys. J. 84:3190–3196, 2003.
Parke, C. L., E. J. Wojcik, S. Kim and D. K. Worthylake. ATP hydrolysis in eg5 kinesin involves a catalytic two-water mechanism. J. Biol. Chem. 285:5859–5867, 2010.
Phillips J. C., R. Braun, W. Wang, J. Gumbart, E. Tajkhorshid, et al. Scalable molecular dynamics with namd. J. Comput. Chem. 26:1781–1802, 2005.
Reubold, T. F., S. Eschenburg, A. Becker, F. J. Kull and D. J. Manstein. A structural model for actin-induced nucleotide release in myosin. Nat. Struct. Biol. 10:826–830, 2003.
Rosenfeld, S. S., P. M. Fordyce, G. M. Jefferson, P. H. King and S. M. Block. Stepping and stretching: How kinesin uses internal strain to walk processively. J. Biol. Chem. 278:18550–18556, 2003.
Rosenfeld, S. S., J. Xing, G. M. Jefferson, H. C. Cheung and P. H. King. Measuring kinesin’s first step. J. Biol. Chem. 277:36731–36739, 2002.
Sablin E. P., and R. J. Fletterick. Nucleotide switches in molecular motors: structural analysis of kinesins and myosins. Curr. Opin. Struct. Biol. 11:716–724, 2001.
Sablin, E. P., F. J. Kull, R. Cooke, R. D. Vale and R. J. Fletterick. Crystal structure of the motor domain of the kinesin-related motor ncd. Nature 380:555–559, 1996.
Sack, S., F. J. Kull and E. Mandelkow. Motor proteins of the kinesin family structures, variations, and nucleotide binding sites. Eur. J. Biochem. 262:1–11, 1999.
Shang, Z., K. Zhou, C. Xu, R. Csencsits, J. C. Cochran, et al.. High-resolution structures of kinesin on microtubules provide a basis for nucleotide-gated force-generation. eLife 3:e04686, 2014.
Sindelar, C. A seesaw model for intermolecular gating in the kinesin motor protein. Biophys. Rev. 3:85–100, 2011.
Sindelar C. V., M. J. Budny, S. Rice, N. Naber, R. Fletterick, et al. Two conformations in the human kinesin power stroke defined by x-ray crystallography and epr spectroscopy. Nat. Struct. Mol. Biol. 9:844–848, 2002.
Sindelar, C. V. and K. H. Downing. The beginning of kinesin’s force-generating cycle visualized at 9-Å resolution. J. Cell Biol. 177:377–385, 2007.
Sindelar, C. V. and K. H. Downing. An atomic-level mechanism for activation of the kinesin molecular motors. Proc. Natl. Acad. Sci. USA 107:4111–4116, 2010.
Skiniotis, G., J. C. Cochran, J. Müller, E. Mandelkow, S. P. Gilbert, et al. Modulation of kinesin binding by the C-termini of tubulin. EMBO J. 23:989–999, 2004.
Smith, C. and I. Rayment. Active site comparisons highlight structural similarities between myosin and other p-loop proteins. Biophys. J. 70: 1590–1602, 1996.
Song, H., and S. A. Endow. Decoupling of nucleotide- and microtubule-binding sites in a kinesin mutant. Nature 396:587–590, 1998.
Toprak, E., A. Yildiz, M. T. Hoffman, S. S. Rosenfeld and P. R. Selvin. Why kinesin is so processive. Proc. Natl. Acad. Sci. USA 106:12717–12722, 2009.
Uchimura, S., Y. Oguchi, Y. Hachikubo, S. Ishiwata and E. Muto. Key residues on microtubule responsible for activation of kinesin ATPase. EMBO J. 29:1167–1175, 2010.
Uemura, S. and S. Ishiwata. Loading direction regulates the affinity of ADP for kinesin. Nat. Struct. Mol. Biol. 10:308–311, 2003.
Vale R. D. The molecular motor toolbox for intracellular transport. Cell 112:467–480, 2003.
Vale R. D., and R. A. Milligan. The way things move: Looking under the hood of molecular motor proteins. Science 288:88–95, 2000.
Zhang, Z. and D. Thirumalai. Dissecting the kinematics of the kinesin step. Structure 20:628–640, 2012.
Acknowledgements
This work was supported by the National Natural Science Foundation of China under Grant Nos. 11545014, 11605038 and the Open Project Program of State Key Laboratory of Theoretical Physics, Institute of Theoretical Physics, Chinese Academy of Science, China (Grant No. Y5KF211CJ1).
Conflict of Interest
Yumei Jin, Yizhao Geng, Lina Lü, Yilong Ma, Gang Lü, Hui Zhang, and Qing Ji declare that they have no conflicts of interest.
Ethical Standards
No human studies were carried out by the authors for this article. No animal studies were carried out by the authors for this article.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Associate Editor Mian Long oversaw the review of this article.
Rights and permissions
About this article
Cite this article
Jin, Y., Geng, Y., Lü, L. et al. Anchor Effect of Interactions Between Kinesin’s Nucleotide-Binding Pocket and Microtubule. Cel. Mol. Bioeng. 10, 162–173 (2017). https://doi.org/10.1007/s12195-017-0477-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12195-017-0477-8