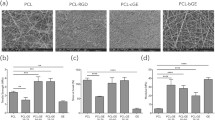
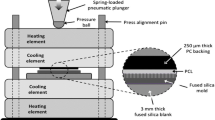
Abstract
Forced expression of transcription factors epigenetically reprograms somatic cells harvested from routine skin biopsies into induced pluripotent stem cells (iPSCs). Human iPSCs are key resources for drug discovery, regenerative medicine and tissue engineering. Here we developed a materials approach to explore how culture substrates could impact factor-mediated reprogramming of human fibroblasts. A materials library consisting of nanofibrous substrates with randomly oriented and aligned structures was prepared by electrospinning four polymers [polylactic acid (PLA), polycaprolactone (PCL), thermoplastic polyurethane (TPU) and polypropylene carbonate (PPC)] into nanofiber orientations. Adsorbing protein to each substrate permitted robust attachment of fibroblasts to all substrates. Fibroblasts on aligned substrates had elongated nuclei, but after reprogramming factor expression, nuclei became more circular. Reprogramming factors could override the nuclear shape constraints imposed by nanofibrous substrates, and the majority of substrates supported full reprogramming. Early culture on PCL and TPU substrates promoted reprogramming, and TGF-β repressed substrate effects. Partial least squares modeling of the biochemical and biophysical cues within our reprogramming system identified TGF-β and polymer identity as important cues governing cellular reprogramming responses. We believe that our approach of using a nanofibrous materials library can be used to dissect molecular mechanisms of reprogramming and generate novel substrates that enhance epigenetic reprogramming.






Similar content being viewed by others
References
Apostolou, E., and K. Hochedlinger. Chromatin dynamics during cellular reprogramming. Nature 502:462–471, 2013.
Armond, J. W., K. Saha, A. A. Rana, C. J. Oates, R. Jaenisch, M. Nicodemi, and S. Mukherjee. A stochastic model dissects cell states in biological transition processes. Sci. Rep. 4:3692, 2014.
Barnes, C. P., S. A. Sell, E. D. Boland, D. G. Simpson, and G. L. Bowlin. Nanofiber technology: designing the next generation of tissue engineering scaffolds. Adv. Drug Deliv. Rev. 59:1413–1433, 2007.
Beers, J., D. R. Gulbranson, N. George, L. I. Siniscalchi, J. Jones, J. A. Thomson, and G. Chen. Passaging and colony expansion of human pluripotent stem cells by enzyme-free dissociation in chemically defined culture conditions. Nat. Protoc. 7:2029–2040, 2012.
Chen, G., Z. Hou, D. R. Gulbranson, and J. A. Thomson. Actin-myosin contractility is responsible for the reduced viability of dissociated human embryonic stem cells. Cell Stem Cell 7:240–248, 2010.
Chen, G., D. R. Gulbranson, Z. Hou, J. M. Bolin, V. Ruotti, M. D. Probasco, K. Smuga-Otto, S. E. Howden, N. R. Diol, N. E. Propson, R. Wagner, G. O. Lee, J. Antosiewicz-Bourget, J. M. Teng, and J. A. Thomson. Chemically defined conditions for human iPSC derivation and culture. Nat. Methods 8:424–429, 2011.
Craene, B. D., and G. Berx. Regulatory networks defining EMT during cancer initiation and progression. Nat. Rev. Cancer 13:97–110, 2013.
de Jong, S. SIMPLS: an alternative approach to partial least squares regression. Chemometr. Intell. Lab. Syst. 18:251–263, 1993.
Discher, D. E., D. J. Mooney, and P. W. Zandstra. Growth factors, matrices, and forces combine and control stem cells. Science 324:1673–1677, 2009.
Downing, T. L., J. Soto, C. Morez, T. Houssin, A. Fritz, F. Yuan, J. Chu, S. Patel, D. V. Schaffer, and S. Li. Biophysical regulation of epigenetic state and cell reprogramming. Nat. Mater. 12:1154–1162, 2013.
Eriksson, L. Multi- and Megavariate Data Analysis, MKS Umetrics AB, 2006.
Feng, B., J.-H. Ng, J.-C. D. Heng, and H.-H. Ng. Molecules that promote or enhance reprogramming of somatic cells to induced pluripotent stem cells. Cell Stem Cell 4:301–312, 2009.
Gaspar-Maia, A., A. Alajem, E. Meshorer, and M. Ramalho-Santos. Open chromatin in pluripotency and reprogramming. Nat. Rev. Mol. Cell Biol. 12:36–47, 2011.
Graf, T., and T. Enver. Forcing cells to change lineages. Nature 462:587–594, 2009.
Grskovic, M., A. Javaherian, B. Strulovici, and G. Q. Daley. Induced pluripotent stem cells—opportunities for disease modelling and drug discovery. Nat. Rev. Drug Discov. 10:915–929, 2011.
Hanna, J. H., K. Saha, and R. Jaenisch. Pluripotency and cellular reprogramming: facts, hypotheses, unresolved issues. Cell 143:508–525, 2010.
Hanna, J., K. Saha, B. Pando, J. van Zon, C. J. Lengner, M. P. Creyghton, A. van Oudenaarden, and R. Jaenisch. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature 462:595–601, 2009.
Haynes, J., J. Srivastava, N. Madson, T. Wittmann, and D. L. Barber. Dynamic actin remodeling during epithelial–mesenchymal transition depends on increased moesin expression. Mol. Biol. Cell 22:4750–4764, 2011.
Hockemeyer, D., F. Soldner, E. G. Cook, Q. Gao, M. Mitalipova, and R. Jaenisch. A drug-inducible system for direct reprogramming of human somatic cells to pluripotency. Cell Stem Cell 3:346–353, 2008.
Jain, N., K. V. Iyer, A. Kumar, and G. V. Shivashankar. Cell geometric constraints induce modular gene-expression patterns via redistribution of HDAC3 regulated by actomyosin contractility. PNAS 110:11349–11354, 2013.
Jiao, J., Y. Dang, Y. Yang, R. Gao, Y. Zhang, Z. Kou, X. F. Sun, and S. Gao. Promoting reprogramming by FGF2 reveals that the extracellular matrix is a barrier for reprogramming fibroblasts to pluripotency. Stem Cells 31:729–740, 2012.
Kim, I. L., S. Khetan, B. M. Baker, C. S. Chen, and J. A. Burdick. Fibrous hyaluronic acid hydrogels that direct MSC chondrogenesis through mechanical and adhesive cues. Biomaterials 34:5571–5580, 2013.
Kohen, N. T., L. E. Little, and K. E. Healy. Characterization of Matrigel interfaces during defined human embryonic stem cell culture. Biointerphases 4:69–79, 2009.
Li, R., J. Liang, S. Ni, T. Zhou, X. Qing, H. Li, W. He, J. Chen, F. Li, Q. Zhuang, B. Qin, J. Xu, W. Li, J. Yang, Y. Gan, D. Qin, S. Feng, H. Song, D. Yang, B. Zhang, L. Zeng, L. Lai, M. A. Esteban, and D. Pei. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell 7:51–63, 2010.
Liu, X., H. Sun, J. Qi, L. Wang, S. He, J. Liu, C. Feng, C. Chen, W. Li, Y. Guo, D. Qin, G. Pan, J. Chen, D. Pei, and H. Zheng. Sequential introduction of reprogramming factors reveals a time-sensitive requirement for individual factors and a sequential EMT–MET mechanism for optimal reprogramming. Nat. Cell Biol. 15:829–838, 2013.
Mali, P., L. Yang, K. M. Esvelt, J. Aach, M. Guell, J. E. DiCarlo, J. E. Norville, and G. M. Church. RNA-guided human genome engineering via Cas9. Science 339:823–826, 2013.
Mattout, A., A. Biran, and E. Meshorer. Global epigenetic changes during somatic cell reprogramming to iPS cells. J. Mol. Cell Biol. 3:341–350, 2011.
McNulty, J. D., T. Klann, J. Sha, M. Salick, G. T. Knight, L. S. Turng, and R. S. Ashton. High-precision robotic microcontact printing (R-μCP) utilizing a vision guided selectively compliant articulated robotic arm. Lab Chip 14:1923–1930, 2014.
Mi, H.-Y., X. Jing, B. R. Jacques, L.-S. Turng, and X.-F. Peng. Characterization and properties of electrospun thermoplastic polyurethane blend fibers: effect of solution rheological properties on fiber formation. J. Mater. Res. 28:2339–2350, 2013.
Ohgushi, M., M. Matsumura, M. Eiraku, K. Murakami, T. Aramaki, A. Nishiyama, K. Muguruma, T. Nakano, H. Suga, M. Ueno, T. Ishizaki, H. Suemori, S. Narumiya, H. Niwa, and Y. Sasai. Molecular pathway and cell state responsible for dissociation-induced apoptosis in human pluripotent stem cells. Cell Stem Cell 7:225–239, 2010.
Quintanilla, R. H., Jr., J. S. T. Asprer, C. Vaz, V. Tanavde, and U. Lakshmipathy. CD44 is a negative cell surface marker for pluripotent stem cell identification during human fibroblast reprogramming. PLoS ONE 9:e85419, 2014.
Rais, Y., A. Zviran, S. Geula, O. Gafni, E. Chomsky, S. Viukov, A. A. Mansour, I. Caspi, V. Krupalnik, M. Zerbib, I. Maza, N. Mor, D. Baran, L. Weinberger, D. A. Jaitin, D. Lara-Astiaso, R. Blecher-Gonen, Z. Shipony, Z. Mukamel, T. Hagai, S. Gilad, D. Amann-Zalcenstein, A. Tanay, I. Amit, N. Novershtern, and J. H. Hanna. Deterministic direct reprogramming of somatic cells to pluripotency. Nature 502:65–70, 2013.
Rnjak-Kovacina, J., S. G. Wise, Z. Li, P. K. Maitz, C. J. Young, Y. Wang, and A. S. Weiss. Tailoring the porosity and pore size of electrospun synthetic human elastin scaffolds for dermal tissue engineering. Biomaterials 32:6729–6736, 2011.
Saha, K., and R. Jaenisch. Technical challenges in using human induced pluripotent stem cells to model disease. Cell Stem Cell 5:584–595, 2009.
Saha, K., Y. Mei, C. M. Reisterer, N. K. Pyzocha, J. Yang, J. Muffat, M. C. Davies, M. R. Alexander, R. Langer, D. G. Anderson, and R. Jaenisch. Surface-engineered substrates for improved human pluripotent stem cell culture under fully defined conditions. PNAS 108:18714–18719, 2011.
Samavarchi-Tehrani, P., A. Golipour, L. David, H. K. Sung, T. A. Beyer, A. Datti, K. Woltjen, A. Nagy, J. L. Wrana. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell 7:64–77, 2010.
Schindelin, J., I. Arganda-Carreras, E. Frise, V. Kaynig, M. Longair, T. Pietzsch, S. Preibisch, C. Rueden, S. Saalfeld, B. Schmid, J. Y. Tinevez, D. J. White, V. Hartenstein, K. Eliceiri, P. Tomancak, and A. Cardona. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9:676–682, 2012.
Shahbazian, M. D., and M. Grunstein. Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 76:75–100, 2007.
Shinagawa, T., T. Takagi, D. Tsukamoto, C. Tomaru, L. M. Huynh, P. Sivaraman, T. Kumarevel, K. Inoue, R. Nakato, Y. Katou, T. Sado, S. Takahashi, A. Ogura, K. Shirahige, and S. Ishii. Histone variants enriched in oocytes enhance reprogramming to induced pluripotent stem cells. Cell Stem Cell 14:217–227, 2014.
Takahashi, K., and S. Yamanaka. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126:663–676, 2006.
Takahashi, K., and S. Yamanaka. Induced pluripotent stem cells in medicine and biology. Development 140:2457–2461, 2013.
Teo, W., and S. Ramakrishna. A review on electrospinning design and nanofibre assemblies. Nanotechnology 17:R89, 2006.
Wang, X., M. R. Salick, X. Wang, T. Cordie, W. Han, Y. Peng, Q. Li, and L. S. Turng. Poly (ε-caprolactone) nanofibers with a self-induced nanohybrid Shish–Kebab structure mimicking collagen fibrils. Biomacromolecules 14:3557–3569, 2013.
Watanabe, K., M. Ueno, D. Kamiya, A. Nishiyama, M. Matsumura, T. Wataya, J. B. Takahashi, S. Nishikawa, S. Nishikawa, K. Muguruma, and Y. Sasai. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat. Biotechnol. 25:681–686, 2007.
Yu, J., M. A. Vodyanik, K. Smuga-Otto, J. Antosiewicz-Bourget, J. L. Frane, S. Tian, J. Nie, G. A. Jonsdottir, V. Ruotti, R. Stewart, I. I. Slukvin, and J. A. Thomson. Induced pluripotent stem cell lines derived from human somatic cells. Science 318:1917–1920, 2007.
Acknowledgments
We acknowledge generous financial support from the Wisconsin Institute for Discovery (T.C. and K.S.), a Grainger Fellowship (J.C-S.) and the Society in Science Foundation (K.S.). We also would like to thank all members of the Saha lab and BIONATES theme for advice and support throughout this project. We acknowledge Dr. Rob McClain and the University of Wisconsin-Madison Biochemistry department for the use and expertise in collecting surface area data via the BET Micromeritics Gemini VII instrument.
Conflict of interest
Travis Cordie, Ty Harkness, Xin Jing, Jared Carlson-Stevermer, Hao-Yang Mi, Lih-Sheng Turng and Krishanu Saha declare that they have no conflicts of interest. Dr. Saha reports grants from Society in Science Foundation during the conduct of the study.
Ethical Standards
No human and animal studies were carried out by the authors for this article. All work with human embryonic stem cell lines was carried out in accordance with institutional, national, and international guidelines and approved by the Stem Cell Research Oversight Committee at the University of Wisconsin-Madison.
Author information
Authors and Affiliations
Corresponding author
Additional information
Associate Editor David Mooney oversaw the review of this article.
This paper is part of the 2014 Young Innovators Issue.
Travis Cordie and Ty Harkness have contributed equally to this work.
Krishanu Saha is an Assistant Professor in the Department of Biomedical Engineering at the University of Wisconsin-Madison. He is also a member of the Wisconsin Institute for Discovery in the bionanocomposite tissue engineering scaffolds (BIONATES) theme. Prior to his arrival in Madison, Dr. Saha studied Chemical Engineering at Cornell University and at the University of California in Berkeley. In his dissertation with Professors David Schaffer and Kevin Healy, he worked on experimental and computational analyses of neural stem cell development, as well as the design of new materials for adult stem cell culture. In 2009 he became a Society in Science: Branco-Weiss fellow in the laboratory of Professor Rudolf Jaenisch at the Whitehead Institute for Biomedical Research at MIT and in the Science and Technology Studies program at Harvard University with Professor Sheila Jasanoff in Cambridge, Massachusetts. Since then, he has performed research on human pluripotent stem cells, disease modeling and synthetic biology.

Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Cordie, T., Harkness, T., Jing, X. et al. Nanofibrous Electrospun Polymers for Reprogramming Human Cells. Cel. Mol. Bioeng. 7, 379–393 (2014). https://doi.org/10.1007/s12195-014-0341-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12195-014-0341-z