Abstract
Protein folding and disaggregation are crucial processes for survival of cells under unfavorable conditions. A network of molecular chaperones supports these processes. Collaborative action of Hsp70 and Hsp100 proteins is an important component of this network. J-proteins/DnaJ members as co-chaperones assist Hsp70. As against 22 DnaJ sequences noted in yeast, rice genome contains 104 J-genes. Rice J-genes were systematically classified into type A (12 sequences), type B (9 sequences), and type C (83 sequences) classes and a scheme of nomenclature of these proteins is proposed. Transcript expression profiles revealed that J-proteins are possibly involved in basal cellular activities, developmental programs, and in stress. Ydj1 is the most abundant J-protein in yeast. Ydj1 deleted yeast cells are nonviable at 37 °C. Two rice ortholog proteins of yeast Ydj1 protein namely OsDjA4 and OsDjA5 successfully rescued the growth defect in mutant yeast. As Hsp70 and J-proteins work in conjunction, it emerges that rice J-proteins can partner with yeast Hsp70 proteins in functioning. It is thus shown that J-protein machine is highly conserved.



Similar content being viewed by others
References
Cajo GC, Horne BE, Kelley WL, Schwager F, Georgopoulos C, Genevaux P (2006) The role of the DIF motif of the DnaJ (Hsp40) co-chaperone in the regulation of the DnaK (Hsp70) chaperone cycle. J Biol Chem 281:12436–12444
Caplan AJ, Cyr DM, Douglas MG (1992) YDJ1p facilitates polypeptide translocation across different intracellular membranes by a conserved mechanism. Cell 71:1143–1155
Chapple JP, van der Spuy J, Poopalasundaram S, Cheetham ME (2004) Neuronal DnaJ proteins HSJ1a and HSJ1b: a role in linking the Hsp70 chaperone machine to the ubiquitin–proteasome system? Biochem Soc Trans 32:640–642
Cyr DM, Lu X, Douglas MG (1992) Regulation of Hsp70 function by a eukaryotic DnaJ homolog. J Biol Chem 267:20927–20931
Dorn KV, Willmund F, Schwarz C, Henselmann C, Pohl T, Hess B, Veyel D, Usadel B, Friedrich T, Nickelsen J, Schroda M (2010) Chloroplast DnaJ-like proteins 3 and 4 (CDJ3/4) from Chlamydomonas reinhardtii contain redox-active Fe–S clusters and interact with stromal HSP70B. Biochem J 427:205–215
Glover JR, Lindquist S (1998) Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell 94:73–82
Griessl MH, Jungkunz I, Sonnewald U, Muller YA (2012) Purification, crystallization and preliminary X-ray diffraction analysis of the Hsp40 protein CPIP1 from Nicotiana tabacum. Acta Crystallogr F Struct Biol Crystallogr Commun 68:236–239
Han W, Christen P (2003) Mechanism of the targeting action of DnaJ in the DnaK molecular chaperone system. J Biol Chem 278:19038–19043
Hennessy F, Cheetham ME, Dirr HW, Blatch GL (2000) Analysis of the levels of conservation of the J domain among the various types of DnaJ-like proteins. Cell Stress Chaperones 5:347–358
Hennessy F, Nicoll WS, Zimmermann R, Cheetham ME, Blatch GL (2005) Not all J domains are created equal: implications for the specificity of Hsp40–Hsp70 interactions. Protein Sci 14:1697–1709
Hofius D, Maier AT, Dietrich C, Jungkunz I, Bornke F, Maiss E, Sonnewald U (2007) Capsid protein-mediated recruitment of host DnaJ-like proteins is required for Potato virus Y infection in tobacco plants. J Virol 81:11870–11880
Kampinga HH, Craig EA (2010) The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol 11:579–592
Kelley WL (1999) Molecular chaperones: how J domains turn on Hsp70s. Curr Biol 9:R305–R308
Kluck CJ, Patzelt H, Genevaux P, Brehmer D, Rist W, Schneider-Mergener J, Bukau B, Mayer MP (2002) Structure–function analysis of HscC, the Escherichia coli member of a novel subfamily of specialized Hsp70 chaperones. J Biol Chem 277:41060–41069
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948
Liu C, Willmund F, Whitelegge JP, Hawat S, Knapp B, Lodha M, Schroda M (2005) J-domain protein CDJ2 and HSP70B are a plastidic chaperone pair that interacts with vesicle-inducing protein in plastids 1. Mol Biol Cell 16:1165–1177
Lu Z, Cyr DM (1998) The conserved carboxyl terminus and zinc finger-like domain of the co-chaperone Ydj1 assist Hsp70 in protein folding. J Biol Chem 273:5970–5978
Lu L, Du Z, Qin M, Wang P, Lan H, Niu X, Jia D, Xie L, Lin Q, Wu Z (2009) Pc4, a putative movement protein of Rice stripe virus, interacts with a type I DnaJ protein and a small Hsp of rice. Virus Genes 38:320–327
Mayer MP, Bukau B (2005) Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol Life Sci 62:670–684
Miernyk JA (2001) The J-domain proteins of Arabidopsis thaliana: an unexpectedly large and diverse family of chaperones. Cell Stress Chaperones 6:209–218
Miot M, Reidy M, Doyle SM, Hoskins JR, Johnston DM, Genest O, Vitery MC, Masison DC, Wickner S (2011) Species-specific collaboration of heat shock proteins (Hsp) 70 and 100 in thermotolerance and protein disaggregation. Proc Natl Acad Sci U S A 108:6915–6920
Misselwitz B, Staeck O, Rapoport TA (1998) J proteins catalytically activate Hsp70 molecules to trap a wide range of peptide sequences. Mol Cell 2:593–603
Mumberg D, Muller R, Funk M (1995) Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156:119–122
Nicoll WS, Botha M, McNamara C, Schlange M, Pesce ER, Boshoff A, Ludewig MH, Zimmermann R, Cheetham ME, Chapple JP, Blatch GL (2007) Cytosolic and ER J-domains of mammalian and parasitic origin can functionally interact with DnaK. Int J Biochem Cell Biol 39:736–751
Ohtsuka K, Hata M (2000) Mammalian HSP40/DNAJ homologs: cloning of novel cDNAs and a proposal for their classification and nomenclature. Cell Stress Chaperones 5:98–112
Qi Y, Wang H, Zou Y, Liu C, Liu Y, Wang Y, Zhang W (2011) Over-expression of mitochondrial heat shock protein 70 suppresses programmed cell death in rice. FEBS Lett 585:231–239
Qiu XB, Shao YM, Miao S, Wang L (2006) The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell Mol Life Sci 63:2560–2570
Rajan VB, D’Silva P (2009) Arabidopsis thaliana J-class heat shock proteins: cellular stress sensors. Funct Integr Genomics 9:433–446
Sahi C, Craig EA (2007) Network of general and specialty J protein chaperones of the yeast cytosol. Proc Natl Acad Sci U S A 104:7163–7168
Shen L, Kang YG, Liu L, Yu H (2011) The J-domain protein J3 mediates the integration of flowering signals in Arabidopsis. Plant Cell 23:499–514
Sielaff B, Tsai FT (2010) The M-domain controls Hsp104 protein remodeling activity in an Hsp70/Hsp40-dependent manner. J Mol Biol 402:30–37
Suetsugu N, Takano A, Kohda D, Wada M (2010) Structure and activity of JAC1 J-domain implicate the involvement of the cochaperone activity with HSC70 in chloroplast photorelocation movement. Plant Signal Behav 5:1602–1606
Sung DY, Guy CL (2003) Physiological and molecular assessment of altered expression of Hsc70-1 in Arabidopsis. Evidence for pleiotropic consequences. Plant Physiol 132:979–987
Takano A, Suetsugu N, Wada M, Kohda D (2010) Crystallographic and functional analyses of J-domain of JAC1 essential for chloroplast photorelocation movement in Arabidopsis thaliana. Plant Cell Physiol 51:1372–1376
Terada K, Oike Y (2010) Multiple molecules of Hsc70 and a dimer of DjA1 independently bind to an unfolded protein. J Biol Chem 285:16789–16797
Valencia-Morales MD, Camas-Reyes JA, Cabrera-Ponce JL, Alvarez-Venegas R (2012) The Arabidopsis thaliana SET-domain-containing protein ASHH1/SDG26 interacts with itself and with distinct histone lysine methyltransferases. J Plant Res 125(5):679–692
Vembar SS, Jin Y, Brodsky JL, Hendershot LM (2009) The mammalian Hsp40 ERdj3 requires its Hsp70 interaction and substrate-binding properties to complement various yeast Hsp40-dependent functions. J Biol Chem 284:32462–32471
Walsh P, Bursac D, Law YC, Cyr D, Lithgow T (2004) The J-protein family: modulating protein assembly, disassembly and translocation. EMBO Rep 5:567–571
Yamamoto T, Mori Y, Ishibashi T, Uchiyama Y, Ueda T, Ando T, Hashimoto J, Kimura S, Sakaguchi K (2005) Interaction between proliferating cell nuclear antigen (PCNA) and a DnaJ induced by DNA damage. J Plant Res 118:91–97
Yamamoto M, Maruyama D, Endo T, Nishikawa S (2008) Arabidopsis thaliana has a set of J proteins in the endoplasmic reticulum that are conserved from yeast to animals and plants. Plant Cell Physiol 49:1547–1562
Yang KZ, Xia C, Liu XL, Dou XY, Wang W, Chen LQ, Zhang XQ, Xie LF, He L, Ma X, Ye D (2009) A mutation in Thermosensitive Male Sterile 1, encoding a heat shock protein with DnaJ and PDI domains, leads to thermosensitive gametophytic male sterility in Arabidopsis. Plant J 57:870–882
Zhou W, Zhou T, Li MX, Zhao CL, Jia N, Wang XX, Sun YZ, Li GL, Xu M, Zhou RG, Li B (2012) The Arabidopsis J-protein AtDjB1 facilitates thermotolerance by protecting cells against heat-induced oxidative damage. New Phytol 194:364–378
Acknowledgement
We thank Jeffrey L. Brodsky of the University of Pittsburgh, USA for DnaJ yeast mutants and Hao Yu of the National University of Singapore, Singapore for kindly providing the seeds of T-DNA insertion mutants of Arabidopsis J3 protein (j3-1 and j3-2). We thank Centre for Plant Molecular Biology and Indo-Finland project, Department of Biotechnology, Govt. of India, for financial support.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary information
Below is the link to the electronic supplementary material.
ESM Fig. 1
FGENESH analysis of Os03g12236 (PDF 27 kb)
ESM Fig. 2
Multiple sequence alignment of J-domain of rice J-proteins showing consensus sequences and four helices characteristic of J-domain. The sequences were aligned in Clustal and edited in Jalview. The consensus sequence and the Logo were derived using Jalview. The amino acid residues are colored by default color scheme of ClustalX. Conserved tripeptide HPD is present between helix II and III. J-proteins in which HPD tripeptide was absent are depicted in red color font (EPS 4,581 kb)
ESM Fig. 3
Tandem arrangement of J-proteins on chromosome (PPTX 387 kb)
ESM Fig. 4
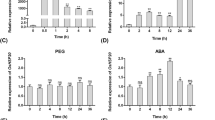
Microarray based expression meta-analysis of type C J-genes in abiotic stresses (a), types C and D during development stages of rice plant (b) and in various tissues (c) (PPTX 346 kb)
ESM Fig. 5
Microarray based expression meta-analysis of types A and B J-genes in development stages (a) and various tissues of rice plant (b) (PPTX 175 kb)
ESM Table 1
Prediction of localization of J-proteins of rice (XLSX 16 kb)
ESM Table 2
List of primers used in this study (XLSX 10 kb)
ESM Table 3
Details of J-proteins of rice without HPD tripeptide in J-domain (XLSX 12.2 kb)
Rights and permissions
About this article
Cite this article
Sarkar, N.K., Thapar, U., Kundnani, P. et al. Functional relevance of J-protein family of rice (Oryza sativa). Cell Stress and Chaperones 18, 321–331 (2013). https://doi.org/10.1007/s12192-012-0384-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12192-012-0384-9