Abstract
Lignin reduction through breeding and genetic modification has the potential to reduce costs in biomass processing in pulp and paper, forage, and lignocellulosic ethanol industries. Here, we present detailed characterization of the extractability and lignin structure of Eucalyptus urophylla × Eucalyptus grandis RNAi downregulated in p-coumaroyl quinate/shikimate 3′-hydroxylase (C3′H) or cinnamate-4-hydroxylase (C4H). Both the C3′H and C4H downregulated lines were found to have significantly higher extractability when exposed to NaOH base extraction, indicating altered cell wall construction. The molecular weight of isolated lignin was measured and lignin structure was determined by HSQC NMR-based lignin subunit analysis for control and the C3′H and C4H downregulated lines. The slight reductions in average molecular weights of the lignin isolated from the transgenic lines (C3′H = 7000, C4H = 6500, control = 7300) does not appear to explain the difference in extractability. The HSQC NMR-based lignin subunit analysis showed increases in H lignin content for the C3′H but only slight differences in the lignin subunit structure of the C3′H and C4H downregulated lines when compared to the control. The greatest difference between the C3′H and C4H downregulated lines is the total lignin content; therefore, it appears that overall lowered lignin content contributes greatly to reduced recalcitrance and increased extractability of cell wall biopolymers. Further studies will be conducted to determine how the reduction in lignin content creates a less rigid cell wall that is more prone to extraction and sugar release.
Similar content being viewed by others
Introduction
Lignocellulose is a complex interlinking structure composed of cellulose, hemicelluloses, and lignin making up the bulk of terrestrial biomass. Commercial processing of lignocellulosic material for paper and biofuels is an energy-intensive process due to the recalcitrant nature of lignocellulose to release cellulose [1–4]. Understanding and overcoming the recalcitrant nature of biomass has been an area of research in recent years. Lignin has been shown to play a major role in biomass processing where both total lignin content and lignin monomer composition may impact the ease with which biomass is broken down to sugars and fermented [3–6].
Three monomers polymerize to form the majority of the lignin polymer: 4-coumaryl alcohol (H), coniferyl alcohol (G), and sinapyl alcohol (S) (Fig. 1) [7, 8]. The proportion of each unit depends on the species, age, cell type, and tissue type [9, 10]. The presence of each additional methoxy group on a lignin unit results in one less reactive site (S < G < H) and, therefore, fewer possible combinations during the polymerization reaction. As a result, lignin rich in S subunits can depolymerize more easily than lignin rich in G subunits as there are fewer C-C bonds and a greater proportion of the more readily cleaved β-O-4 linkages. In addition to the core S/G/H polymer, lignin includes a range of other aromatic structures commonly bonded to the main lignin structure through ether or ester linkages. Such modifications (especially frequent in herbaceous species) usually involve the addition of p-coumaric acid, ferulic acid, and occasionally, sinapic acid. The modifications can take the form of cross-linking groups between lignin and carbohydrate [11–13], or pendant groups at the γ-position of the lignin interlinkage unit [14–17].
The C3′H and C4H genes are both involved in the production of S and G lignin monomers in the lignin biosynthetic pathway. Downregulation of C3′H is expected to reduce production of S and G monomers and has been shown to lead to an increase in incorporation of H monomers in lignin for many plant species [18–23]. While this result was observed in alfalfa [18] and maize [19], herbaceous species make a greater proportion of H lignin naturally [20], so the outcome for tree species may be different. C3′H downregulation was first achieved in Arabidopsis when Franke et al. [21, 22] isolated the REF8 gene and demonstrated that it encodes a functional C3′H enzyme. The Arabidopsis ref8 mutant contains approximately half as much lignin as the wild type and was the first known mutant line published to have lignin almost entirely comprised of H lignin subunits, with only traces of S and G. A transgenic T-DNA insertion mutant of the CYP98A3 gene, which is the gene affected by the ref8 mutant, also had reduced lignin and primarily H lignin subunits [23]. Coleman et al. [24] published work on C3′H downregulation of a hybrid poplar, Populus grandidentata × alba. Their work demonstrated total lignin yields ranged from equivalent to the WT, to less than 50 % (line C3H-14) of the WT. S/G ratio remained equivalent to the WT (1.8), except for the most severely downregulated line (C3H-14), where S/G ratio increased to 3.1. This occurred as a result of an increase in H subunits of lignin (WT = 0.2 %, C3H-14 = 20.6 %) at the expense of G content (WT = 35.5 %, C3H-14 = 19.3 %) [24].
In the present work, transgenic hybrid eucalyptus (E. urophyll × E. grandis) lines downregulated in p-coumaroyl quinate/shikimate 3′-hydroxylase (C3′H) or cinnamate-4-hydroxylase (C4H) is used as a model to understand the role of C3′H and C4H in lignin polymerization for tree species. Previous work published studying these C3′H and C4H downregulated eucalyptus samples demonstrated that both C3′H and C4H downregulated sets of lines had lowered average S/G ratio (C3′H = 1.2, C4H = 1.2, control = 2.1) and lignin content (C3′H = 21.0 %, C4H = 21.7 %, control = 30.4 %) [25]. The lines also had large decreases in the average recalcitrance as assessed by sugar release after hydrothermal pretreatment and enzymatic saccharification (C3′H = 93 %, C4H = 95 %, control = 80 %). Thioacidolysis results indicated an increase in H lignin for only the C3′H line, with an average of 18.3 % H lignin, compared to the C4H lines at 1.2 % and the control with 1.0 %. This increase in H lignin for only the C3′H lines is consistent with its locations in the lignin biosynthetic pathway where C3′H is only involved in the synthesis of S and G monomers but C4H is located at a point that downregulates all the monomers, including H [24].
To better understand the changes in lignin in these transgenic E. urophylla × grandis lines, we utilize sequential extraction to chemically dissect the cell wall material. As highlighted in Ziebell et al. [18], sequential extraction is a powerful tool for analyzing the impact of lignin pathway modifications on the plant cell wall. Progressively, more aggressive treatments are used to remove decreasingly labile molecules from the lignocellulosic matrix. Prior solvent extraction ensures that simple metabolites do not interfere with the interpretation of the cell wall chemistry results. In addition to the extractability profiles, molecular weight profiles and lignin subunit analysis have been performed on the samples to determine how the chemical composition of the lignin polymer has been altered due to the C3′H and C4H downregulation.
Material and Methods
Materials
Wood with reduced lignin from transgenic hybrid eucalyptus (E. urophylla × E. grandis) was provided by ArborGen Inc. The same wood material was used in this work as in Sykes et al. [25]. The plants were grown for 3 months as containerized cuttings in the greenhouse, acclimatized for 1 month outdoors, and then transferred into the field in Glades County, Florida, for an additional 20 months before destructive sampling. A stem section from an individual tree for each line was debarked, air dried, and milled with a Wiley mill to pass through a 20-mesh screen to produce approximately 20 g of material that was utilized for the work in both Sykes et al. [25] and the work presented here. Two representative lines were selected from each of the C3′H reduced, C4H reduced, and control sets previously characterized in Sykes et al. [25]. Methanol (HPLC grade), tetrahydrofuran (THF, HPLC grade), glacial acetic acid (reagent grade), and diethyl ether (anhydrous, reagent grade) were from JT Baker, Phillipsburg, NJ. The 1,4-dioxane (anhydrous, 99.80 %), 1,2-dichloroethane (≥99.8 % HPLC grade), acetic anhydride (reagent grade), and pyridine (anhydrous, ≥99.8 %) were sourced from Sigma-Aldrich, MO, USA.
Sequential Extraction
Air dried, ground (∼0.5 g, 60 mesh) wood was weighed into teabags and extracted with a 32:68 toluene to ethanol solution for 48 h in a Soxhlet Extractor. The solids from the toluene to ethanol extraction (0.350 g) were weighed into 60-mL vials, 0.1 M NaOH (25 mL) was added and the samples were shaken at 5 °C overnight prior to filtration. The vials were washed (4 × 2.5 mL H2O) as was the solid residue (4 × 2.5 mL, H2O) and the washes and filtrates were collected. The crucibles were washed with water until the water wash was neutral (∼45 mL, not collected). The solid was freeze dried and weighed.
An aliquot of each sample was further extracted with 2.0 M NaOH. Approximately 200 mg of the 0.1 M NaOH extracted material was added to 60-mL vials and 25 mL of 2.0 M NaOH was added. The samples were stirred in a 110 °C oil bath for 10 min. Samples were cooled in a water bath at room temperature. The filtrate was acidified to pH 2.0 with concentrated hydrochloric acid (HCl). The solution was allowed to rest at 4 °C overnight before the precipitate was filtered off, freeze dried, and weighed.
Lignin Isolation
Ball-milled lignin was isolated from extractive-free eucalyptus according to modified literature methods described previously [26, 27]. Wiley milled biomass samples (60 mesh) were Soxhlet extracted with toluene to ethanol: (32:68, v/v) for 36 h and then dried under vacuum overnight. The extractive-free biomass (5.0-10.0 g) was ball-milled in a Roalox ceramic jar (0.3 L) (U.S. Stoneware, Ohio, USA) with zirconia balls using a rotary ball mill running at 96 rpm for 8 days under N2 atm to prevent oxidation. A zirconia ball to biomass weight ratio of 32:1 was used. The ball-milled residue was then extracted (2 × 48-h stirring, in the dark) with 1,4-dioxane-water (96:4, v/v) (10 mL/g biomass). The extracted mixture was centrifuged, and the supernatant was collected and evaporated at 35 °C under reduced pressure to obtain the crude ball-milled lignin. The crude lignin was dissolved in acetic acid-water (9:1, v/v) (50 mg/mL) and precipitated into de-ionized water (200 mL), centrifuged, washed with de-ionized water (2 × 20 ml), and dried under vacuum at room temperature for 24 h. For further purification, the solid product was dissolved in a minimum quantity of 1,2-dichloroethane to ethanol (2:1, v/v) and precipitated into diethyl ether (200 mL), centrifuged, washed with diethyl ether (3 × 20 ml), dried under vacuum (40 °C, overnight) and stored in a refrigerator. The ball-milled lignin yield based on lignin content in extracted eucalyptus was 7.27 % for control b, 6.80 % for control a, 8.12 % for C3′H 105, 8.35 % for C3′H 106b, 5.12 % for C4H 73a, and 5.21 % for C4H 99a.
Gel Permeation Chromatography
The isolated lignin samples were weighed into Reacti-vials (5 mL, Thermo Scientific) and acetylated by dissolving in pyridine (10 μL/1 mg sample), stirring at 40 °C for 1 h, adding acetic anhydride (10 μL/1 mg sample) and stirring overnight at room temperature. The reaction was terminated by addition of methanol (4 μL/1 mg sample). The acetylation reagents were evaporated from the samples at 40 °C under a stream of nitrogen gas, and the samples were then dried in a vacuum oven at 40 °C overnight, then in a vacuum desiccator (1 Torr) at room temperature for 1 h. The dried acetylated samples were dissolved in THF and filtered (0.45-μm nylon membrane syringe filters) before GPC analysis.
GPC analysis was performed using an Agilent HPLC with three polystyrene-divinyl benzene GPC columns (Polymer Laboratories, 300 × 7.5 mm, 10-μm beads) having nominal pore diameters of 104, 103, and 102 Å. The eluent was THF, the flow rate was 1.0 mL/min, the sample concentration was ∼2 mg/mL, and an injection volume of 25 μL was used. The HPLC was attached to a diode array detector measuring absorbance at 260 nm (band width 40 nm). Polystyrene calibration standards were used with molecular weights ranging from 580 Da to 2.95 million Da. Toluene was used as the monomer calibration standard.
Solid-State NMR
Solid-state NMR spectra were collected on the extracted cell wall residues using high-resolution 13C cross-polarization/magic angle spinning (CP/MAS) with a Bruker Avance 200 MHz spectrometer (50.13 MHz, room temperature). The spinning speed was 7000 Hz and a contact time of 2 ms with a 1-dB ramp on the proton spin locking field was applied during cross polarization. The acquisition time was 32.8 ms and the recycle delay was 1 s.
HSQC NMR
Heteronuclear Single Quantum Coherence (HSQC) NMR spectra were acquired for isolated ball-milled lignin samples (10 mg), dissolved in DMSO-d6 (500 μL). Spectra were acquired at 40 °C on a Bruker Avance III 600 MHz spectrometer at 11.7 T using a TCI cryoprobe. Spectra were acquired with 1024 points and a SW of 15 ppm in the F2 (1H) dimension and 512 points and SW of 220 ppm in the F1 (13C) dimension. For the lignin subunit analysis, integrations were performed on the peak representative of the α C-H correlation for A (β-O-4), B (β-β resinol), C (β-5 phenylcoumaran), and D (β-1 spirodienone) subunits, while the γ C-H is used for F (cinnamyl alcohol end groups). The spectral processing parameters from Mansfield et al. [28] were used and integrations were performed using TopSpin 3.2. Designations and assignments for these groups are given in Kim et al. [29] and Rencoret et al. [30].
Results and Discussion
Sequential Extraction
Solvent was first used to remove all small metabolites to prevent them from interfering with later analysis. The removal of extractives is important for samples such as eucalyptus, which have high extractive content due to the natural oils in the plant. All samples showed a loss of material on extraction with toluene-ethanol with the variation between lines in the same transformation type being equivalent to the variation across all the samples. Due to this lack of differentiation between the control and the transgenic lines on toluene-ethanol extraction, the extractives were not further investigated.
More material was extracted from the transgenic lines than the control lines during both base extraction steps as shown in Fig. 2. During the 0.1 M NaOH extraction, twice as much material was extracted from the transgenic lines (C3′H = 12.9 %, C4H = 10.8 %) compared to the control (5.4 %). While the 2.0 M NaOH extraction removed approximately 50 % more material from the transgenic lines (C3′H = 37.8 %, C4H = 37.3 %) than the control (24.4 %). The extractability profile in Fig. 2 demonstrates that the downregulated lines are significantly altered from the control. The increase in extractability has also been seen in alfalfa [18, 31] and indicates that material in the plant cell wall is less tightly held in these transgenic lines downregulated in lignin. However, the exact changes to the three-dimensional structure of the plant cell wall are not known from this analysis. The largest difference in extractability between the transgenic lines and the control was seen on extraction with 0.1 M NaOH. Under these conditions the compounds removed are loosely bound, base labile components of the plant cell wall, which includes phenolics and a significant portion of the hemicelluloses [18]. Analysis of the resultant extracted biomass by solid-state 13C NMR gives more information about the residual biomass and, therefore, further explores the differences between the control and the transgenic lines.
Solid-State NMR of the Cell Wall Residue
Solid-state 13C cross polarization magic angle spinning (13C CP/MAS) NMR spectroscopy was used to investigate the lignin structural characteristics in the plant cell walls. The advantage of this technique over liquid-state NMR is the inclusion of the whole biomass instead of only the soluble components. Solid-state 13C NMR allows a greater proportion of the biomass to be examined, although it does have lower resolution. Six samples, two C3′H, two C4H, and two controls were selected for these studies and the replicates within each gene construct demonstrated good reproducibility. For figure clarity and ease of reading, only the control b, C3′H 105, and C4H 73a lines are presented in NMR Figs. 3, 4, 5, and 6. Figures with data for control a, C3′H 106b, and C4H 99a are presented in the supplementary materials.
The treatment of biomass with strong base removes both phenolic and hemicellulosic components of the plant cell wall, making the before and after spectra potentially difficult to compare. However, as demonstrated previously in Ziebell et al. [18], the C4 carbon of the crystalline cellulose peak (89 ppm) [32–34] can be used as an internal standard when mild treatments are applied and the crystalline cellulose peak in the NMR spectra remains a consistent width and shape. By adjusting the solid-state NMR spectra of the three lines so they are equal at 89 ppm (assigned to the C4 carbon in interior crystalline cellulose), the amount of lignin relative to crystalline cellulose in each sample can be compared.
Comparison of the solid-state 13C NMR of the transgenic lines with the control in Fig. 3 shows that there is a greater proportion of aromatic material (110–165 ppm) in the control, indicating a higher lignin content, consistent with the previously published pyrolysis molecular beam mass spectrometry (py-MBMS) data [25]. The aromatic region of the extracted plant cell wall residue is usually assigned to lignin, although conjugated carbons in lignin side chains or other lignin moieties can appear in this region cross-linked to either lignin or hemicellulose fractions (phenolic compounds such as ferulates and coumarates). The region also represents tannins and tannic compounds. More specifically, the higher control lignin content can be seen from the prominence of peaks 153 ppm (S3/5) [35] and 133–8 ppm (S1/4 and G1) [36] in the aromatic region compared to the transgenic lines (Fig. 3). In addition, lower methoxyl content in the transgenic lines is indicated by the reduction of the methoxyl peak at 56 ppm which indicates a concomitant reduction in the lignin methoxyl group. A decrease in methoxyl content can also result from a change in H:G:S content of lignin as each move from a S to a G lignin subunit, or from a G to a H subunit, represents a loss of one methoxyl group. It is impossible from these results, however, to know what portion of the aromatic material is actual lignin from solely the solid-state 13C NMR. Comparison with Klason lignin and py-MBMS results from Sykes et al. [25] agree with this trend in the NMR data. One notable difference between the C3′H and C4H lines is the presence of a shoulder in the C3′H lines between 120 and 130 ppm (Fig. 3) that is assigned to H2 and H6 positions in H subunit of lignin. For the C4H lines there is also loss of signal intensity in the region 110–122 ppm, compared to the C3′H and control lines, which corresponds to both G (G2 = 112, G5 = 116, and G6 = 120 ppm) and H (H3/5 = 114–118 [37] or 121–123 ppm [38]) subunits. The C4H line also displays greater intensity in the carbohydrate peaks between 60–80 ppm that is representative of the higher glucose levels in the C4H lines as shown in Sykes et al. [25].
The solid-state 13C NMR representation of the sequential extraction of the control and transgenic lines is shown in Fig. 4. On treatment with 0.1 M NaOH, the C3′H lines show loss of material in the aromatic portion of the NMR spectrum, and all samples show a decrease in of a large proportion of the acetyl peaks (166–176 ppm). The largest loses of aromatic material on 0.1 M NaOH extraction of the C3′H lines corresponds to the S3/5 peak at 153 ppm and the S1/4/G1 peak at 133–8 ppm. Although the solid-state NMR spectra of the C3′H lines displayed a decrease in the aromatic region of the spectra after base treatment, it seems unlikely that a dilute base could remove true lignin and it is more likely that the loss represents structures similar to S and G subunits of lignin (therefore appearing in the same region of the spectrum) but not polymerized into the actual lignin polymer. These could be hydroxycinnamic acids and/or modified lignin which is more easily extracted than lignin from unmodified biomass. Modified lignin was identified in Ziebell et al. [18] and the base extractives from the present study demonstrated low solubility after neutralization, consistent with modified lignin. In fact, analysis on the extracted materials was not possible because of the lack of solubility, which is an unlikely problem for the analysis of hydroxycinnamic acids at low concentration in water.
The C4H lines do not have a significant peak in the 124–131 ppm region (H lignin), but they also show little change in intensity in this region on extraction with dilute or concentrated base. The fact that the extraction profiles vary between the C3′H and C4H transgenic lines, highlights the difference in downregulation at the two different points in the biosynthetic pathway (C3′H vs. C4H). Much less aromatic material is removed from the C4H lines during base extraction and, although greatly decreased in intensity (because of the lowered lignin content), the distribution of the material within the aromatic region of the NMR spectrum does not change as much when compared to the control and the C3′H lines. The presence of increased H subunits in the solid-state NMR spectra of the C3′H lines is consistent with the previously published thioacidolysis results [25] that show the control having 1.0 % H subunits, the C4H lines having 1.2 % H subunits and the C3′H line having 18.3 % H subunits. The lack of any distinct H4 subunit peak at 158 ppm in the C4H lines means that the signal loss in the region 110–122 ppm can mostly likely be attributed to loss of G subunits as H subunits do not appear to be present to any large extent. This is consistent with the lower S/G ratio of the C4H lines (py-MBMS), and in having H subunit content similar to the controls.
For the 2.0 M NaOH extractions, reductions in the 125–155 ppm region of the spectra are observed when compared to the 0.1 M NaOH extraction for both the C4H and C3′H lines while the control spectra is relatively unchanged. Although harsh enough to remove all residual hemicelluloses, this treatment should not remove fully intact native lignin. Therefore, the reduction in this region indicates that the transformed lines have a portion of lignin or lignin-like compounds that are not bound into the native lignin structure and are more easily extracted from the cell wall. For the C3′H line there is a reduction in the signal intensity of the 110–130 ppm region that corresponds to the region containing primarily H lignin. For all the samples, the acetyl region (166–176 ppm) of the NMR spectra is completely removed by the strong, 2.0 M base extraction.
Gel Permeation Chromatography
Gel permeation chromatography (GPC) was performed on isolated ball-milled lignin preparations for the six samples studied in this paper (Table 1 and Fig. 5). Only small changes in the molecular weight profiles of the isolated lignins were seen. The molecular weight profiles of the isolated lignin of C3′H 105 differs more from the control than C3′H 106b; however, the difference is slight. C3′H 105 has the lowest number averaged molecular weight (Mn) and weight-averaged molecular weight (Mw) at 3140 Da and 6670 Da respectively. This is compared to C3′H 106b (Mn = 3310 Da and Mw = 7280 Da), control b (Mn = 3340 Da Mw = 7100 Da) and control a (Mn = 3480 Da and Mw = 7430 Da). There is a greater difference between the two C4H lines with C4H 73a having similar Mw and Mn to the controls (Mn = 3420 Da and Mw = 6710 Da) and C4H 99a being much lower (Mn = 2870 Da and Mw = 6290 Da). The standard deviations of the two controls were calculated and assumed to be representative of the natural variation between untransformed plants (Mn(sdWT) = 99 Da and Mw(sdWT) = 233 Da). Taking into account a WT variation of two standard deviations, the isolated lignin of lines C3′H 105 and C4H 99a are statistically different to the averaged control result on the basis of Mn and Mw; however, C3′H 106b and C4H 73a are not statistically different. However, care should be taken with this interpretation given that the sample size was only two samples for the molecular weight analysis of the WT and C3ˈH and C4H downregulations. The difference between the four transgenic lines and the two controls can be seen on the GPC plot in Fig. 5 with the difference in the two transgenic lines being slight, but observable.
The small reduction in the molecular weight of lignin in this study is in contrast to more significant changes seen in the molecular weight of isolated lignin in C3H and HCT downregulated alfalfa [18]. When the same estimate of isolated lignin molecular weight was used by Ziebell et al. [18] for C3H downregulated alfalfa, decreases of 15–60 % (Mn) and 10–50 % (Mw) were seen (decreases were even higher for the HCT lines). Given similar decreases in both lignin concentration and recalcitrance in both alfalfa and eucalyptus downregulated in C3′H expression, it begs the question: why were there not similar changes in molecular weight? Alfalfa and eucalyptus are very different species (herbaceous vs. hardwood), and additionally the lignin subunit (H:G:S) distribution is very different. The alfalfa samples downregulated in C3H have very high concentrations of the H lignin subunit (30–55 %) [18, 39], whereas the eucalyptus has much lower H lignin content (18 % average from thioacidolysis [25]). Increasing H lignin subunit concentration has already been suggested as a cause of reduction in molecular weight, with changed chemical-physical properties suggesting an altered reaction tendency compared to G and S lignin subunit types [18].
HSQC NMR
To investigate the structural subunits present in the isolated ball-milled lignin, 2D 1H-13C HSQC NMR of the lignin dissolved in DMSO-d6 were conducted and the spectra are displayed in Fig. 6. The most notable difference in the lines from these spectra is the presence of H lignin in the C3′H line in contrast to both the control and C4H lines, which contain very small amounts, consistent with the thioacidolysis [25] and solid-state NMR results. A lignin subunit analysis was performed by integrating peaks in the HSQC NMR spectra for the relevant subunit. The peaks for each subunit region are labeled in Fig. 6 and the subunit analysis with the relative percentages of each of the units are shown in Table 2. It should be noted that while HSQC NMR spectra are not quantitative, these numbers can serve as a comparison of relative changes between the controls and downregulated lines. For the C3′H downregulated lines there is an increase in C (β-5 phenylcoumaran) linkages from to 7.8 % (average) compared to the control at 5.3 % (average). These linkages can only be formed with G or H type lignin so this increase may be a result of the increased H content in the C3′H lines. There is also an increase in F (cinnamyl alcohol end groups) for the C3′H lines having 5.7 % compared to 3.5 % for the control. The increases in the C and F subunit structures are balanced by a slight decreases in the A (β-O-4) and D (β-1 spirodienone) subunit structures. For the C4H lines the only difference in the subunit structures is an increase in A (β-O-4) linkages with a simultaneous decrease in B (β-β resinol). The S/G ratio and relative S:G:H percentages were calculated based on the HSQC NMR spectra and are also shown in Table 2. The results of the integration of the spectra agree well with the thioacidolysis results with the C3′H lines having an average of 18.3 % H lignin. In addition, the trend in S/G ratios calculated from these results, 1.9 for controls, 1.3 for C3′H, and 1.3 for C4H, also matches well with py-MBMS and thioacidolysis results.
This lignin subunit analysis for the C3′H lines has similar trends in H lignin bonding as previously observed in C3′H downregulated alfalfa [39]. Although the differences were larger in alfalfa, most likely due to the higher H lignin content, the C3′H eucalyptus lines also displayed a decrease in β-O-4 and β-1 subunits and an increase in β-5 subunits. These results are also supported by Density Functional Theory (DFT) simulations that calculated lower reaction enthalpies for β-5 coupling in H lignin dimers [40]. The DFT simulations also calculated lower reaction enthalpies of B (β-β resinol) subunits for S lignin dimers [40]. The work presented here shows a decrease in the β-β resinol units, consistent with the decrease in S/G ratio and the lower amount of S lignin in the C4H lines.
Conclusions
In previous studies on alfalfa, it was hypothesized that H lignin content and lower lignin molecular weights were, at least in part, the drivers for the increased extractability of the lignin-like material. In this work, sequential extractions of both the C3′H and C4H downregulated eucalyptus lines displayed increased extractives content with 0.1 M and 2 M NaOH treatments compared to the control, indicating an altered cell wall. Solid-state 13C NMR of the cell wall residues and HSQC NMR of isolated lignin displayed peaks characteristic of H lignin for the C3′H lines only. The C3′H downregulated lines contain H lignin and C4H lines do not; however, both displayed increased extractability. Therefore, it appears that H lignin content is not the primary cause of increased extractability and reduced recalcitrance in these modified lines. In addition, downregulation of C3′H and C4H did not lead to lower molecular weights in the milled-wood lignin molecular weight profile. These results demonstrate that overall lowered lignin content is the greatest contributor to reduced recalcitrance and increased extractability of cell wall biopolymers. It is hypothesized that the reduction of the lignin content creates a less rigid cell wall that is more prone to extraction and sugar release, and further studies are underway to elucidate the mechanism.
References
Chiang VL (2006) Monolignol biosynthesis and genetic engineering of lignin in trees, a review. Environ Chem Lett 4:143–146
O’Connell A, Holt K, Piquemal J, Grima-Pettenati J, Boudet A, Pollet B et al (2002) Improved paper pulp from plants with suppressed cinnamoyl-CoA reductase or cinnamyl alcohol dehydrogenase. Transgenic Res 11:495
Reddy MSS, Chen F, Shadle G, Jackson L, Aljoe H, Dixon RA (2005) Targeted down-regulation of cytochrome P450 enzymes for forage quality improvement in alfalfa (Medicago sativa L.). Proc Natl Acad Sci U S A 102(46):16573–16578
Chen F, Dixon RA (2007) Lignin modification improves fermentable sugar yields for biofuel production. Nat Biotechnol 25(7):759–761
Shen H, Poovaiah CR, Ziebell A, Tschaplinski TJ, Pattathil S, Gjersing E et al (2013) Enhanced characteristics of genetically modified switchgrass (Panicum virgatum L.) for high biofuel production. Biotechnol Biofuels 6:71
Mansfield SD, Kang KY, Chapple C (2012) Designed for deconstruction— poplar trees altered in cell wall lignification improve the efficacy of bioethanol production. New Phytol 194(1):91–101
Sarkanen KV (1971) Precursors and their polymerization. In: Sarkanen KV, Ludwig CH, editors. Lignins: Occurrence, formation, structure and reactions. Wiley-Interscience
Freudenberg K, Neish AC (1968) Constitution and biosynthesis of lignin. Springer, Berlin
Whetten RW, MacKay JJ, Sederoff RR (1998) Recent advances in understanding lignin biosynthesis. Annu Rev Plant Physiol Plant Mol Biol 49:585–609
Monties B, Fukushima K (2001) Occurance, function and biosynthesis of lignins. In: Hofrichter M, Steinbuchel A (eds) Lignin, humic substances and Coal. Wiley-VCH, Weinheim
Lam TBT, Iiyama K, Stone BA (1992) Cinnamic acid bridges between cell-wall polymers in wheat and phalaris internodes. Phytochemistry 31(4):1179–1183
Iiyama K, Lam TBT, Kasuya N, Stone BA (1994) Rapid and simple determination of o-acetyl groups bound to plant-cell walls by acid-hydrolysis and H-1-NMR measurement. Phytochemistry 35(4):959–961
Iiyama K, Lam TBT, Stone BA (1990) Phenolic-acid bridges between polysaccarides and lignin in wheat internodes. Phytochemistry 29(3):733–737
Buranov AU, Mazza G (2008) Lignin in straw of herbaceous crops. Ind Crop Prod 28(3):237–259
Crestini C, Argyropoulos DS (1997) Structural analysis of wheat straw lignin by quantitative P-31 and 2D NMR spectroscopy. The occurrence of ester bonds and alpha-O-4 substructures. J Agric Food Chem 45(4):1212–1219
Ralph J, Hatfield RD, Quideau S, Helm RF, Grabber JH, Jung HJG (1994) Pathway of p-coumaric acid incorporation into maize lignin as revealed by NMR. J Am Chem Soc 116(21):9448–9456
Ralph J (1996) An unusual lignin from kenaf (1996). J Nat Prod 59(4):341–342. doi:10.1021/np960143s
Ziebell A, Gracom K, Katahira R, Chen F, Pu YQ, Ragauskas A et al (2010) Increase in 4-coumaryl alcohol units during lignification in alfalfa (Medicago sativa) alters the extractability and molecular weight of lignin. J Biol Chem 285(50):38961–38968
Fornale S, Rencoret J, Garcia-Calvo L, Capellades M, Encina A, Santiago R et al (2015) Cell wall modifications triggered by the down-regulation of coumarate 3-hydroxylase-1 in maize. Plant Sci 236:272–282
Bonawitz ND, Chapple C (2010) The genetics of lignin biosynthesis: connecting genotype to phenotype. Annu Rev Genet 44:337–364
Franke R, Hemm M, Denault J, Ruegger M, Humphreys J, Chapple C (2002) Changes in secondary metabolism and deposition of an unusual lignin in the ref8 mutant b nm. Plant J 30(1):47–59
Franke R, Humphreys J, Hemm M, Denault J, Ruegger M, Cusumano J et al (2002) The Arabidopsis REF8 gene encodes the 3-hydroxylase of phenylpropanoid metabolism. Plant J 30(1):33–45
Abdulrazzak N, Pollet B, Ehlting J, Larsen K, Asnaghi C, Ronseau S et al (2006) A coumaroyl-ester-3-hydroxylase insertion mutant reveals the existence of nonredundant meta-hydroylation pathways and essential roles for phenolic precursors in cell expansion and plant growth. Plant Physiol 140:18
Coleman HD, Park J-Y, Nair R, Chapple C, Mansfield SD (2008) RNAi-mediated suppression of p-coumaroyl-CoA 3′-hydroxylase in hybrid poplar impacts lignin deposition and soluble secondary metabolism. Proc Natl Acad Sci U S A 105(11):4501–4506
Sykes R, Gjersing E, Foutz K, Rottmann W, Kuhn S, Foster C et al (2015) Down regulation of p-coumarate 3′-hydroxylase (C3′H) and cinnamate 4-hydroxylase (C4H) genes in the lignin biosynthetic pathway of Eucalyptus urophylla x Eucalyptus grandis leads to improved sugar release. Biotechnol Biofuels 8:128
Pu YQ, Chen F, Ziebell A, Davison BH, Ragauskas AJ (2009) NMR characterization of C3H and HCT down-regulated alfalfa lignin. BioEnergy Res 2(4):198–208
Chang HM, Cowling EB, Brown W, Adler E, Miksche G (1975) Comparative studies on cellulolytic enzyme lignin and milled wood lignin of sweetgum and spruce. Holzforschung 29(5):153–159
Mansfield SD, Kim H, Lu F, Ralph J (2012) Whole plant cell wall characterization using solution-state 2D NMR. Nat Protoc 7(9):1579–1589
Kim H, Ralph J, Akiyama T (2008) Solution-state 2D NMR of ball-milled plant cell wall gels in DMSO-d6. BioEnergy Res 1(1):56–6666
Rencoret J, Marques G, Gutierrez A, Nieto L, Santos JI, Jimenez-Barbero J et al (2008) HSQC-NMR analysis of lignin in woody (Eucalyptus globulus and Picea abies) and non-woody (Agave sisalana) ball-milled plant materials at the gel state 10(th) EWLP, Stockholm, Sweden, August 25–28, 2008. Holzforschung 63(6):691–698
Gallego-Giraldo L, Jikumaru Y, Kamiya Y, Tang Y, Dixon RA (2011) Selective lignin downregulation leads to constitutive defense response expression in alfalfa (Medicago sativa L.). New Phytol 190(3):627–639
Teeäär R, Serimaa R, Paakkarl T (1987) Crystallinity of cellulose, as determined by CP/MAS NMR and XRD methods. Polym Bull 17(3):231–237
Park S, Johnson DK, Ishizawa CI, Parilla PA, Davis MF (2009) Measuring the crystallinity index of cellulose by solid state C-13 nuclear magnetic resonance. Cellulose 16(4):641–647
Hult EL, Liitia T, Maunu SL, Hortling B, Iversen T (2002) A CP/MAS C-13-NMR study of cellulose structure on the surface of refined kraft pulp fibers. Carbohydr Polym 49(2):231–234
Robert D (ed) (1992) Methods in lignin chemistry. Springer, New York
Manders WF (1987) Solid-state C-13 NMR determination of the syringyl guaiacyl ratio in hardwoods. Holzforschung 41(1):13–18
NMR Database of Lignin and Cell Wall Model Compounds (2004) Available from: http://ars.usda.gov/Services/docs.htm?docid=10491. Accessed 9 Mar 2013
Wagner A, Ralph J, Akiyama T, Flint H, Phillips L, Torr K et al (2007) Exploring lignification in conifers by silencing hydroxycinnamoyl-CoA: shikimate hydroxycinnamoyltransferase in Pinus radiata. Proc Natl Acad Sci U S A 104(28):11856
Ralph J, Akiyama T, Kim H, Lu FC, Schatz PF, Marita JM et al (2006) Effects of coumarate 3-hydroxylase down-regulation on lignin structure. J Biol Chem 281(13):8843–8853
Sangha AK, Parks JM, Standaert RF, Ziebell A, Davis M, Smith JC (2012) Radical coupling reactions in lignin synthesis: a density functional theory study. J Phys Chem B 116(16):4760–4768
Acknowledgments
The authors would like to thank William Rottmann, ArborGen Inc., for manuscript review and for providing the biomass materials studied in this paper. The BESC is a U.S. Department of Energy Bioenergy Research Center supported by the Office of Biological and Environmental Research in the DOE Office of Science. This work was supported by the U.S. Department of Energy under Contract No. DE-AC36-08-GO28308 with the National Renewable Energy Laboratory.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 479 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Ziebell, A., Gjersing, E., Hinchee, M. et al. Downregulation of p-Coumaroyl Quinate/Shikimate 3′-Hydroxylase (C3′H) or Cinnamate-4-hydrolylase (C4H) in Eucalyptus urophylla × Eucalyptus grandis Leads to Increased Extractability. Bioenerg. Res. 9, 691–699 (2016). https://doi.org/10.1007/s12155-016-9713-7
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12155-016-9713-7



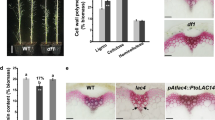



 ) and C3′H down-regulated E. urophylla × E. grandis C3′H 105 (
) and C3′H down-regulated E. urophylla × E. grandis C3′H 105 ( ) and C4H 73a (
) and C4H 73a ( ) extracted with toluene to ethanol
) extracted with toluene to ethanol
 ) and, Toluene:Ethanol (
) and, Toluene:Ethanol ( ), 0.1 M NaOH (
), 0.1 M NaOH ( ) and 2.0 M NaOH (
) and 2.0 M NaOH ( ) extracted biomass
) extracted biomass
