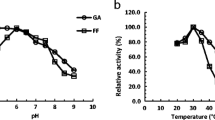
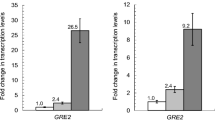
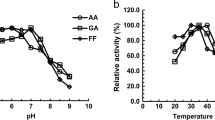
Abstract
Furfural is one of the main aldehyde inhibitors generated during the pretreatment of lignocellulosic biomass. Saccharomyces cerevisiae can in situ detoxify furfural to the less toxic furan methanol (FM) via the activities of multiple dehydrogenases/reductases. In this study, we report that an uncharacterized gene, YML131W, was highly induced under furfural stress conditions and that the transcription factors Yap1p, Msn2/4p, and/or Hsf1p likely controlled its upregulated expression. The induced transcription of YML131W led to higher concentrations of its encoded protein. Enzyme activity assays showed that YML131W is an aldehyde reductase that plays a role in detoxifying furfural to FM. YML131W also showed activity toward other aldehydes, suggesting that it is involved in detoxifying endogenous toxic aldehydes generated via the degradation of misfolded and damaged proteins. This detoxification role would help to maintain cell viability under furfural stress conditions. A S. cerevisiae strain overexpressing YML131W showed increased tolerance to furfural. YML131W was able to catalyze the conversion of formaldehye, acetaldehyde, propionaldehyde, and butyaldehyde to their corresponding alcohols, indicating that it has potential applications in producing fuels such as butanol and isobutanol. A phylogenetic analysis grouped YML131W into the leukotriene B4 dehydrogenases (LTD) family, but its amino acid sequence substantially differed from those of other proteins in the LTD family. We identified 15 proteins from 14 yeast species that showed sequence similarities to YML131W. These other proteins likely have similar functions to that of YML131W and may have potential to confer tolerance to aldehyde inhibitors derived from the lignocellulosic biomass conversion process.








Similar content being viewed by others
References
Lin Y, Tanaka S (2006) Ethanol fermentation from biomass resources: current state and prospects. Appl Microbiol Biotechnol 69:627–642
Nielsen J, Larsson C, van Maris A, Pronk J (2013) Metabolic engineering of yeast for production of fuels and chemicals. Curr Opin Biotechnol 24:398–404
Delgenes JP, Moletta R, Navarro JM (1996) Effects of lignocellulose degradation products on ethanol fermentations of glucose and xylose by Saccharomyces cerevisiae, Zymomonas mobilis, Pichias tipitis, and Candida shehatae. Enzym Microb Technol 19:220–225
Larsson S, Palmqvist E, Hahn-Hägerdal B, Tengborg C, Stenberg K, Zacchi G, Nilvebrant NO (1999) The generation of inhibitors during dilute acid hydrolysis of softwood. Enzym Microb Technol 24:151–159
Luo CD, Brink DL, Blanch HW (2002) Identification of potential fermentation inhibitors in conversion of hybrid poplar hydrolyzate to ethanol. Biomass Bioenerg 22:125–138
Liu ZL, Blaschek HP (2010) Biomass conversion inhibitors and in situ detoxification. In: Vertès AA, Qureshi N, Blaschek HP, Yukawa H (eds) Biomass to biofuels: strategies for global industries. Wiley, Chichester, pp 233–259
Antal MJ, Leesomboon T, Mok WS, Richards GN (1991) Mechanism of formation of 2-furaldehyde from D-xylose. Carbohydr Res 217:71–85
Taherzadeh MJ, Eklund R, Gustafsson L, Niklasson C, Lidén G (1997) Characterization and fermentation of dilute-acid hydrolyzates from wood. Ind Eng Chem Res 36:4659–4665
Heer D, Sauer U (2008) Identification of furfural as a key toxin in lignocellulosic hydrolysates and evolution of a tolerant yeast strain. Microb Biotechnol 1:497–506
Jönsson LJ, Alriksson B, Nilvebrant NO (2013) Bioconversion of lignocellulose: inhibitors and detoxification. Biotechnol Biofuels 6:16
Liu ZL (2011) Molecular mechanisms of yeast tolerance and in situ detoxification of lignocellulose hydrolysates. Appl Microbiol Biotechnol 90:809–825
Heer D, Heine D, Sauer U (2009) Resistance of Saccharomyces cerevisiae to high concentrations of furfural is based on NADPH-dependent reduction by at least two oxireductases. Appl Environ Microbiol 75:7631–7638
Laadan B, Almeida JR, Rådström P, Hahn-Hägerdal B, Gorwa-Grauslund M (2008) Identification of an NADH-dependent 5-hydroxymethylfurfural-reducing alcohol dehydrogenase in Saccharomyces cerevisiae. Yeast 25:191–198
Liu ZL, Moon J, Andersh BJ, Slininger PJ, Weber S (2008) Multiple gene-mediated NAD(P)H-dependent aldehyde reduction is a mechanism of in situ detoxification of furfural and 5-hydroxymethylfurfural by Saccharomyces cerevisiae. Appl Microbiol Biotechnol 81:743–753
Liu ZL, Moon J (2009) A novel NADPH-dependent aldehyde reductase gene from Saccharomyces cerevisiae NRRL Y-12632 involved in the detoxification of aldehyde inhibitors derived from lignocellulosic biomass conversion. Gene 446:1–10
Park SE, Koo HM, Park YK, Park SM, Park JC, Lee OK, Park YC, Seo JH (2011) Expression of aldehyde dehydrogenase 6 reduces inhibitory effect of furan derivatives on cell growth and ethanol production in Saccharomyces cerevisiae. Bioresour Technol 102:6033–6038
Moon J, Liu ZL (2012) Engineered NADH-dependent GRE2 from Saccharomyces cerevisiae by directed enzyme evolution enhances HMF reduction using additional cofactor NADPH. Enzym Microb Technol 50:115–120
Liu ZL, Slininger PJ, Dien BS, Berhow MA, Kurtzman CP, Gorsich SW (2004) Adaptive response of yeasts to furfural and 5-hydroxymethylfurfural and new chemical evidence for HMF conversion to 2, 5-bis-hydroxymethylfuran. J Ind Microbiol Biotechnol 31:345–352
Liu ZL, Slininger PJ, Gorsich SW (2005) Enhanced biotransformation of furfural and 5-hydroxymethylfurfural by newly developed ethanologenic yeast strains. Appl Biochem Biotechnol 121–124:451–460
Liu ZL, Ma M, Song M (2009) Evolutionarily engineered ethanologenic yeast detoxifies lignocellulosic biomass conversion inhibitors by reprogrammed pathways. Mol Genet Genomics 282:233–244
Sehnem NT, Machado Ada S, Leite FC, Pita Wde B, de Morais MA, Jr AMA (2013) 5-Hydroxymethylfurfural induces ADH7 and ARI1 expression in tolerant industrial Saccharomyces cerevisiae strain P6H9 during bioethanol production. Bioresour Technol 133:190–196
Ma M, Liu ZL (2010) Comparative transcriptome profiling analyses during the lag phase uncover YAP1, PDR1, PDR3, RPN4, and HSF1 as key regulatory genes in genomic adaptation to lignocellulose derived inhibitor HMF for Saccharomyces cerevisiae. BMC Genomics 11:660
Nordling E, Jörnvall H, Persson B (2002) Medium-chain dehydrogenases/reductases (MDR). Family characterizations including genome comparisons and active site modeling. Eur J Biochem 269:4267–4276
Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK (2003) Global analysis of protein localization in budding yeast. Nature 425:686–691
Ma M, Wang X, Zhang X, Zhao X (2013) Alcohol dehydrogenases from Scheffersomyces stipitis involved in the detoxification of aldehyde inhibitors derived from lignocellulosic biomass conversion. Appl Microbiol Biotechnol 97:8411–8425
Rozen S, Skaletsky H (2000) Bioinformatics methods and protocols. In: Krawetz S, Misener S (eds) Methods in molecular biology. Humana, Totowa, pp 365–386
Liu ZL, Slininger PJ (2007) Universal external RNA controls for microbial gene expression analysis using microarray and qRT-PCR. J Microbiol Methods 68:486–496
Liu ZL, Palmquist DE, Ma M, Liu J, Alexander NJ (2009) Application of a master equation for quantitative mRNA analysis using qRT-PCR. J Biotechnol 143:10–16
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Gietz RD, Schiestl RH, Willems AR, Woods RA (1995) Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11:355–360
Teixeira MC, Monteiro P, Jain P, Tenreiro S, Fernandes AR, Mira NP, Alenquer M, Freitas AT, Oliveira AL, Sá-Correia I (2006) The YEASTRACT database: a tool for the analysis of transcription regulatory associations in Saccharomyces cerevisiae. Nucleic Acids Res 34:D446–D451
Hori T, Yokomizo T, Ago H, Sugahara M, Ueno G, Yamamoto M, Kumasaka T, Shimizu T, Miyano M (2004) Structural basis of leukotriene B4 12-hydroxydehydrogenase/ 15-oxo-prostaglandin 13-reductase catalytic mechanism and a possible Src homology 3 domain binding loop. J Biol Chem 279:22615–22623
Marchler-Bauer A, Zheng C, Chitsaz F, Derbyshire MK, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Lu S, Marchler GH, Song JS, Thanki N, Yamashita RA, Zhang D, Bryant SH (2013) CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res 41:D348–D352
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729
Sinicropi D, Cronin M, Liu M-L (2007) Gene expression profiling utilizing microarray technology and RT-PCR. In: Ferrari ME-i-c, Ozkan M, Heller M (eds) BioMEMS and biomedical nanotechnology, Volume II: Micro/Nanotechnologies for genomics and proteomics. Springer-Verlag, Heidelberg, pp 23–46
Alriksson B, Horváth IS, Jönsson LJ (2010) Overexpression of Saccharomyces cerevisiae transcription factor and multidrug resistance genes conveys enhanced resistance to lignocellulose derived fermentation inhibitors. Process Biochem 45:264–271
Sasano Y, Watanabe D, Ukibe K, Inai T, Ohtsu I, Shimoi H, Takagi H (2012) Overexpression of the yeast transcription activator Msn2 confers furfural resistance and increases the initial fermentation rate in ethanol production. J Biosci Bioeng 113:451–455
Kim D, Hahn JS (2013) Roles of the Yap1 transcription factor and antioxidants in Saccharomyces cerevisiae’s tolerance to furfural and 5-hydroxymethylfurfural, which function as thiol-reactive electrophiles generating oxidative stress. Appl Environ Microbiol 79:5069–5077
Persson B, Hedlund J, Jörnvall H (2008) Medium- and short-chain dehydrogenase/reductase gene and protein families: the MDR superfamily. Cell Mol Life Sci 65:3879–3894
Van Dijken JP, Scheffers WA (1986) Redox balances in the metabolism of sugars by yeasts. FEMS Microbiol Rev 32:199–224
Allen SA, Clark W, McCaffery JM, Cai Z, Lanctot A, Slininger PJ, Liu ZL, Gorsich SW (2010) Furfural induces reactive oxygen species accumulation and cellular damage in Saccharomyces cerevisiae. Biotechnol Biofuels 3:2
Goldberg AL (2003) Protein degradation and protection against misfolded or damaged proteins. Nature 426:895–899
Wang X, Xu H, Ha SW, Ju D, Xie Y (2010) Proteasomal degradation of Rpn4 in Saccharomyces cerevisiae is critical for cell viability under stressed conditions. Genetics 184:335–342
Hazelwood LA, Daran JM, van Maris AJ, Pronk JT, Dickinson JR (2008) The Ehrlich pathway for fusel alcohol production: a century of research on Saccharomyces cerevisiae metabolism. Appl Environ Microbiol 74:2259–2266
López-Rituerto E, Avenoza A, Busto JH, Peregrina JM (2010) Evidence of metabolic transformations of amino acids into higher alcohols through 13C NMR studies of wine alcoholic fermentation. J Agric Food Chem 58:4923–4927
Branduardi P, Longo V, Berterame NM, Rossi G, Porro D (2013) A novel pathway to produce butanol and isobutanol in Saccharomyces cerevisiae. Biotechnol Biofuels 6:68
Wonisch W, Schaur RJ, Bilinski T, Esterbauer H (1995) Assessment of growth inhibition by aldehydic lipid peroxidation products and related aldehydes by Saccharomyces cerevisiae. Cell Biochem Funct 13:91–98
Acknowledgments
This work was supported in part by grants from the Science and Technology Department of Sichuan Province (Grant No. 2014HH0013), the Scientific Research Fund of Sichuan Provincial Education Department (Grant No. 13ZB0286), and the Talent Introduction Fund of Sichuan Agricultural University (Grant No. 01426100). We thank Z. Lewis Liu, Bioenergy Research Unit, NCAUR-ARS, US Department of Agriculture (Peoria, IL, USA) for the calibrated mRNA control mix used as the reference mRNA for the qRT-PCR analyses.
Author information
Authors and Affiliations
Corresponding author
Additional information
Xi Li and Ruoheng Yang contributed equally to this paper.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOC 1896 kb)
Rights and permissions
About this article
Cite this article
Li, X., Yang, R., Ma, M. et al. A Novel Aldehyde Reductase Encoded by YML131W from Saccharomyces cerevisiae Confers Tolerance to Furfural Derived from Lignocellulosic Biomass Conversion. Bioenerg. Res. 8, 119–129 (2015). https://doi.org/10.1007/s12155-014-9506-9
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12155-014-9506-9