Abstract
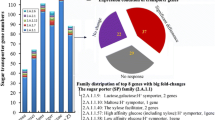
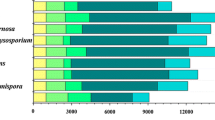
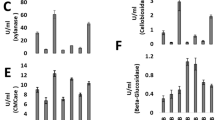
Fusarium oxysporum can convert straw to ethanol via consolidated bioprocessing (CBP)—a two-stage process that firstly involves aerobic saccharification and thereafter an oxygen-limiting fermentation phase. The efficacy of CBP is dependent upon the fungal strain used. Using suppression subtractive hybridisation (SSH), a total of 210 transcripts were identified as being overexpressed in a high as compared to low efficacy CBP strain in the aerobic and oxygen-limiting growth stages on a straw/bran mix. These transcripts encode proteins assigned to various categories, including carbohydrate metabolism, energy, protein and sugar transport and detoxification. Real-time RT-PCR analysis of 12 transcripts, including an endoglucanase III (EGIII), a novel ricin toxin A1 chain-like protein (RTA1), and two unknown transcripts (JX308289 and JX308290) validated the SSH findings. Post-transcriptional silencing of EGIII, RTA1 and an unknown transcript JX308289 in F. oxysporum strain 11C significantly reduced the capacity of the fungus to produce ethanol from a straw/bran mix. Thus these and other genes identified in this study are likely factors that determine the efficacy of CBP and such genes can be used as candidates for enhancing microbial ethanol production from straw and other lignocellulosic substrates.



Similar content being viewed by others
References
Kim S, Dale BE (2004) Global potential bioethanol production from wasted crops and crop residues. Biomass Bioenerg 26(4):361–375
King BC, Waxman KD, Nenni NV, Walker LP, Bergstrom GC, Gibson DM (2011) Arsenal of plant cell wall degrading enzymes reflects host preference among plant pathogenic fungi. Biotechnol Biofuels 4(1):4–18
Anasontzis GE, Zerva A, Stathopoulou PM, Haralampidis K, Diallinas G, Karagouni AD et al (2011) Homologous overexpression of xylanase in Fusarium oxysporum increases ethanol productivity during consolidated bioprocessing (CBP) of lignocellulosics. J Biotechnol 152(1–2):16–23
Fu Z, Holtzapple MT (2010) Consolidated bioprocessing of sugarcane bagasse and chicken manure to ammonium carboxylates by a mixed culture of marine microorganisms. Bioresource Technol 101(8):2825–2836
Xu Q, Singh A, Himmel ME (2009) Perspectives and new directions for the production of bioethanol using consolidated bioprocessing of lignocellulose. Curr Opin Biotechnol 20(3):364–371
Christakopoulos P, Koullas D, Kekos D, Koukios E, Macris B (1991) Direct ethanol conversion of pretreated straw by Fusarium oxysporum. Bioresource Technol 35(3):297–300
Dogaris I, Vakontios G, Kalogeris E, Mamma D, Kekos D (2009) Induction of cellulases and hemicellulases from Neurospora crassa under solid-state cultivation for bioconversion of sorghum bagasse into ethanol. Ind Crops Prod 29(2–3):404–411
Panagiotou G, Christakopoulos P, Olsson L (2005) Simultaneous saccharification and fermentation of cellulose by Fusarium oxysporum F3–growth characteristics and metabolite profiling. Enzyme Microb Technol 36(5–6):693–699
Panagiotou G, Christakopoulos P, Villas-Boas SG, Olsson L (2005) Fermentation performance and intracellular metabolite profiling of Fusarium oxysporum cultivated on a glucose–xylose mixture. Enzyme Microb Technol 36(1):100–106
Ali SS, Khan M, Fagan B, Mullins E, Doohan F (2012) Exploiting the inter-strain divergence of Fusarium oxysporum for microbial bioprocessing of lignocellulose to bioethanol. AMB Express 2(1):16
Diatchenko L, Lau Y, Campbell AP, Chenchik A, Moqadam F, Huang B et al (1996) Suppression subtractive hybridisation: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. P Natl Acad Sci U S A 93(12):6025–6030
Brennan J, Egan D, Cooke B, Doohan F (2005) Effect of temperature on head blight of wheat caused by Fusarium culmorum and F. graminearum. Plant Pathol 54(2):156–160
Mishra C, Keskar S, Rao M (1984) Production and properties of extracellular endoxylanase from Neurospora crassa. Appl Environ Microbiol 48(1):224–228
Chang S, Puryear J, Cairney J (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep 11(2):113–116
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W et al (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25(17):3389–3402
Ansari KI, Walter S, Brennan JM, Lemmens M, Kessans S, McGahern A et al (2007) Retrotransposon and gene activation in wheat in response to mycotoxigenic and non-mycotoxigenic-associated Fusarium stress. Theor Appl Genet 114(5):927–937
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25(4):402–408
Nakayashiki H, Hanada S, Quoc NB, Kadotani N, Tosa Y, Mayama S (2005) RNA silencing as a tool for exploring gene function in ascomycete fungi. Fungal Genet Biol 42(4):275–283
Doohan FM, Smith P, Parry DW, Nicholson P (1998) Transformation of Fusarium culmorum with the [beta]-d-glucuronidase (GUS) reporter gene: a system for studying host–pathogen relationships and disease control. Physiol Mol Plant Pathol 53(5–6):253–268
Scotti CT, Vergoignan C, Feron G, Durand A (2001) Glucosamine measurement as indirect method for biomass estimation of Cunninghamella elegans grown in solid state cultivation conditions. Biochem Eng J 7(1):1–5
Ride JP, Drysdale RB (1972) A rapid method for the chemical estimation of filamentous fungi in plant tissue. Physiol Plant Pathol 2(1):7–15
Snedecor G, Cochran W (1980) Statistical methods, 6th edn. Iowa State University Press, Ames
Rep M, Van Der Does HC, Meijer M, Van Wijk R, Houterman PM, Dekker HL et al (2004) A small, cysteine-rich protein secreted by Fusarium oxysporum during colonization of xylem vessels is required for I-3-mediated resistance in tomato. Mol Microbiol 53(5):1373–1383
Van Der Does HC, Lievens B, Claes L, Houterman PM, Cornelissen BJC, Rep M (2008) The presence of a virulence locus discriminates Fusarium oxysporum isolates causing tomato wilt from other isolates. Environ Microbiol 10(6):1475–1485
Macarron R, Acebal C, Castillon MP, Dominguez J, de la Mata I, Pettersson G et al (1993) Mode of action of endoglucanase III from Trichoderma reesei. Biochem J 289(3):867–873
Ward RJ (2011) In: Buckeridge MS, Goldman GH (eds) Cellulase engineering for biomass saccharification routes to cellulosic ethanol. Springer, New York, pp 135–151
Manente M, Ghislain M (2009) The lipid-translocating exporter family and membrane phospholipid homeostasis in yeast. FEMS Yeast Res 9(5):673–687
Sehgal A, Hughes BT, Espenshade PJ (2008) Oxygen-dependent, alternative promoter controls translation of tco1+ in fission yeast. Nucleic Acids Res 36(6):2024–2031
Kołaczkowska A, Manente M, Kołaczkowski M, Laba J, Ghislain M, Wawrzycka D (2012) The regulatory inputs controlling pleiotropic drug resistance and hypoxic response in yeast converge at the promoter of the aminocholesterol resistance gene RTA1. FEMS Yeast Res 12(3):279–292
Gilead S, Shoham Y (1995) Purification and characterization of alpha-L-arabinofuranosidase from Bacillus stearothermophilus T-6. Appl Environ Microbiol 61(1):170–174
Stephanopoulos G (2007) Challenges in engineering microbes for biofuels production. Science 315(5813):801–804
Cohen JI, Roychowdhury S, DiBello PM, Jacobsen DW, Nagy LE (2009) Exogenous thioredoxin prevents ethanol induced oxidative damage and apoptosis in mouse liver. Hepatology 49(5):1709–1717
Coon M, Ding X, Pernecky S, Vaz A (1992) Cytochrome P450: progress and predictions. FASEB J 6(2):669–673
Leão C, Uden N (1985) Effects of ethanol and other alkanols on the temperature relations of glucose transport and fermentation in Saccharomyces cerevisiae. Appl Microbiol Biotechnol 22(5):359–363
Ali SS, Nugent B, Mullins E, Doohan FM (2013) Insights from the fungus Fusarium oxysporum point to high affinity glucose transporters as targets for enhancing ethanol production from lignocellulose. PLoS One 8(1):e54701
Brandao RL, Loureiro-Dias MC (1990) Regulation of sugar transport systems in Fusarium oxysporum var. lini. Appl Environ Microbiol 56(8):2417–2420
Atsumi S, Hanai T, Liao JC (2008) Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature 451(7174):86–89
Dunham MJ, Badrane H, Ferea T, Adams J, Brown PO, Rosenzweig F et al (2002) Characteristic genome rearrangements in experimental evolution of Saccharomyces cerevisiae. P Natl Acad Sci U S A 99(25):16144–16149
Hughes TR, Roberts CJ, Dai H, Jones AR, Meyer MR, Slade D et al (2000) Widespread aneuploidy revealed by DNA microarray expression profiling. Nat Genet 25(3):333–337
Finnegan DJ (1989) Eukaryotic transposable elements and genome evolution. Trends Genet 5(4):103–107
Hua-Van A, Langin T, Daboussi MJ (2001) Evolutionary history of the impala transposon in Fusarium oxysporum. Mol Biol Evol 18(10):1959–1969
Acknowledgements
This work was supported by the Irish Department of Agriculture, Fisheries and Food Research Stimulus Fund (RSF 07 513).
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 660 kb)
Rights and permissions
About this article
Cite this article
Ali, S.S., Khan, M., Mullins, E. et al. Identification of Fusarium oxysporum Genes Associated with Lignocellulose Bioconversion Competency. Bioenerg. Res. 7, 110–119 (2014). https://doi.org/10.1007/s12155-013-9353-0
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12155-013-9353-0