Abstract
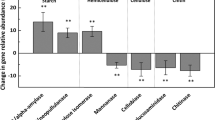
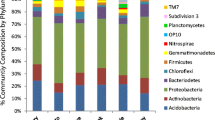
We obtained soil samples from geographically diverse switchgrass (Panicum virgatum L.) and sorghum (Sorghum bicolor L.) crop sites and from nearby reference grasslands and compared their edaphic properties, microbial gene diversity and abundance, and active microbial biomass content. We hypothesized that soils under switchgrass, a perennial, would be more similar to reference grassland soils than sorghum, an annual crop. Sorghum crop soils had significantly higher NO3 − -N, NH4 + -N, SO4 2− -S, and Cu levels than grassland soils. In contrast, few significant differences in soil chemistry were observed between switchgrass crop and grassland soils. Active bacterial biomass was significantly lower in sorghum soils than switchgrass soils. Using GeoChip 4.0 functional gene arrays, we observed that microbial gene diversity was significantly lower in sorghum soils than grassland soils. Gene diversity at sorghum locations was negatively correlated with NO3 − -N, NH4 + -N, and SO4 2− -S in C and N cycling microbial gene categories. Microbial gene diversity at switchgrass sites varied among geographic locations, but crop and grassland sites tended to be similar. Microbial gene abundance did not differ between sorghum crop and grassland soils, but was generally lower in switchgrass crop soils compared to grassland soils. Our results suggest that switchgrass has fewer adverse impacts on microbial soil ecosystem services than cultivation of an annual biofuel crop such as sorghum. Multi-year, multi-disciplinary regional studies comparing these and additional annual and perennial biofuel crop and grassland soils are recommended to help define sustainable crop production and soil ecosystem service practices.




Similar content being viewed by others
References
Ragauskas AJ, Williams CK, Davison BH, Britovsek G, Cairney J, Eckert CA et al (2006) The path forward for biofuels and biomaterials. Sci 311(5760):484–489. doi:10.1126/science.1114736
Fargione J, Hill J, Tilman D, Polasky S, Hawthorne P (2008) Land clearing and the biofuel carbon debt. Sci 319(5867):1235–1238. doi:10.1126/science.1152747
Carroll A, Somerville C (2009) Cellulosic biofuels. Annual Rev Plant Biology 60(1):165–182. doi:10.1146/annurev.arplant.043008.092125
Davis SC, Anderson-Teixeira KJ, DeLucia EH (2009) Life-cycle analysis and the ecology of biofuels. Trends Plant Sci 14(3):140–146
Tilman D, Socolow R, Foley JA, Hill J, Larson E, Lynd L et al (2009) Beneficial biofuels—the food, energy, and environment trilemma. Sci 325(5938):270–271. doi:10.1126/science.1177970
Moon HS, Abercrombie JM, Kausch AP, Stewart CN Jr (2010) Sustainable use of biotechnology for bioenergy feedstocks. Environ Manag 46(4):531–538. doi:10.1007/s00267-010-9503-5
Hill J, Nelson E, Tilman D, Polasky S, Tiffany D (2006) Environmental, economic, and energetic costs and benefits of biodiesel and ethanol biofuels. PNAS 103(30):11206–11210. doi:10.1073/pnas.0604600103
Tilman D, Wedin D, Knops J (1996) Productivity and sustainability influenced by biodiversity in grassland ecosystems. Nat 379(6567):718–720
Hooper DU, Vitousek PM (1997) The effects of plant composition and diversity on ecosystem processes. Sci 277(5330):1302–1305
Tilman D, Hill J, Lehman C (2006) Carbon-negative biofuels from low-input high-diversity grassland biomass. Sci 314(5805):1598–1600. doi:10.1126/science.1133306
Dale VH, Kline KL, Wiens J, Fargione J (2010) Biofuels: implications for land use and biodiversity. Ecological Society of America, Washington, DC
Maestre FT, Quero JL, Gotelli NJ, Escudero A, Ochoa V, Delgado-Baquerizo M et al (2012) Plant species richness and ecosystem multifunctionality in global drylands. Sci 335(6065):214–218. doi:10.1126/science.1215442
Midgley GF (2012) Biodiversity and ecosystem function. Sci 335(6065):174–175. doi:10.1126/science.1217245
Tilman D, Knops J, Wedin D, Reich P, Ritchie M, Siemann E (1997) The influence of functional diversity and composition on ecosystem processes. Sci 277(5330):1300–1302. doi:10.1126/science.277.5330.1300
Dobranic JK, Zak JC (1999) A microtiter plate procedure for evaluating fungal functional diversity. Mycologia 91(5):756–765
Bossio DA, Girvan MS, Verchot L, Bullimore J, Borelli T, Albrecht A et al (2005) Soil microbial community response to land use change in an agricultural landscape of western Kenya. Microb Ecol 49(1):50–62. doi:10.1007/s00248-003-0209-6
He Z, Gentry TJ, Schadt CW, Wu L, Liebich J, Chong SC et al (2007) GeoChip: a comprehensive microarray for investigating biogeochemical, ecological and environmental processes. ISME J 1(1):67–77, http://www.nature.com/ismej/journal/v1/n1/suppinfo/ismej20072s1.html
Yergeau E, Kang S, He Z, Zhou J, Kowalchuk GA (2007) Functional microarray analysis of nitrogen and carbon cycling genes across an Antarctic latitudinal transect. ISME J 1(2):163–179, http://www.nature.com/ismej/journal/v1/n2/suppinfo/ismej200724s1.html
Zhang Y, Zhang X, Liu X, Xiao Y, Qu L, Wu L et al (2007) Microarray-based analysis of changes in diversity of microbial genes involved in organic carbon decomposition following land use/cover changes. FEMS Microbiol Lett 266(2):144–151. doi:10.1111/j.1574-6968.2006.00511.x
Zhou J, Kang S, Schadt CW, Garten CT Jr (2008) Spatial scaling of functional gene diversity across various microbial taxa. PNAS 105(22):7768–7773. doi:10.1073/pnas.0709016105
Berthrong ST, Schadt CW, Piñeiro G, Jackson RB (2009) Afforestation alters the composition of functional genes in soil and biogeochemical processes in South American grasslands. Appl Environ Microbiol 75(19):6240–6248. doi:10.1128/aem.01126-09
Da C Jesus E, Susilawati E, Smith S, Wang Q, Chai B, Farris R et al (2010) Bacterial communities in the rhizosphere of biofuel crops grown on marginal lands as evaluated by 16S rRNA gene pyrosequences. BioEnergy Res 3(1):20–27
Lu Z, Deng Y, Van Nostrand JD, He Z, Voordeckers J, Zhou A et al (2012) Microbial gene functions enriched in the Deepwater Horizon deep-sea oil plume. ISME J 6(2):451–460, http://www.nature.com/ismej/journal/vaop/ncurrent/suppinfo/ismej201191s1.html
DeSantis T, Brodie E, Moberg J, Zubieta I, Piceno Y, Andersen G (2007) High-density universal 16S rRNA Microarray analysis reveals broader diversity than typical clone library when sampling the environment. Microb Ecol 53(3):371–383. doi:10.1007/s00248-006-9134-9
Cole JR, Chai B, Farris RJ, Wang Q, Kulam SA, McGarrell DM et al (2005) The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res 33(suppl 1):D294–D296. doi:10.1093/nar/gki038
Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ et al (2009) The ribosomal database project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37(suppl 1):D141–D145. doi:10.1093/nar/gkn879
McGrath KC, Mondav R, Sintrajaya R, Slattery B, Schmidt S, Schenk PM (2010) Development of an environmental functional gene microarray for soil microbial communities. Appl Environ Microbiol 76(21):7161–7170. doi:10.1128/aem.03108-09
Horneck DA, Hart JM, Topper K, Koepsell B (1989) Methods of soil analysis used in the Soil Testing Laboratory at Oregon State University. Agricultural Experiment Station, Oregon State University, Corvallis
Ingham ER, Klein DA (1984) Soil fungi: relationships between hyphal activity and staining with fluorescein diacetate. Soil Biol Biochem 16(3):273–278. doi:10.1016/0038-0717(84)90014-2
Schnürer J, Rosswall T (1982) Fluorescein diacetate hydrolysis as a measure of total microbial activity in soil and litter. Appl Environ Microbiol 43(6):1256–1261
Van Veen JA, Ladd JN, Frissel MJ (1984) Modelling C and N turnover through the microbial biomass in soil. Plant Soil 76(1):257–274. doi:10.1007/bf02205585
Hazen TC, Dubinsky EA, DeSantis TZ, Andersen GL, Piceno YM, Singh N et al (2010) Deep-sea oil plume enriches indigenous oil-degrading bacteria. Sci 330(6001):204–208. doi:10.1126/science.1195979
He Z, Deng Y, Van Nostrand JD, Tu Q, Xu M, Hemme CL et al (2010) GeoChip 3.0 as a high-throughput tool for analyzing microbial community composition, structure and functional activity. ISME J 4(9):1167–1179, http://www.nature.com/ismej/journal/v4/n9/suppinfo/ismej201046s1.html
Wu L, Liu X, Schadt CW, Zhou J (2006) Microarray-based analysis of subnanogram quantities of microbial community DNAs by using whole-community genome amplification. Appl Environ Microbiol 72(7):4931–4941. doi:10.1128/aem.02738-05
Liang Y, He Z, Wu L, Deng Y, Li G, Zhou J (2010) Development of a common oligonucleotide reference standard for microarray data normalization and comparison across different microbial communities. Appl Environ Microbiol 76(4):1088–1094. doi:10.1128/aem.02749-09
He Z, Xu M, Deng Y, Kang S, Kellogg L, Wu L et al (2010) Metagenomic analysis reveals a marked divergence in the structure of belowground microbial communities at elevated CO2. Ecol Lett 13(5):564–575. doi:10.1111/j.1461-0248.2010.01453.x
Girvan MS, Bullimore J, Pretty JN, Osborn AM, Ball AS (2003) Soil type is the primary determinant of the composition of the total and active bacterial communities in arable soils. Appl Environ Microbiol 69(3):1800–1809. doi:10.1128/aem.69.3.1800-1809.2003
Bezemer TM, Lawson CS, Hedlund K, Edwards AR, Brook AJ, Igual JM et al (2006) Plant species and functional group effects on abiotic and microbial soil properties and plant-soil feedback responses in two grasslands. J Ecol 94(5):893–904. doi:10.1111/j.1365-2745.2006.01158.x
Pereira e Silva MC, Poly F, Guillaumaud N, van Elsas JD, Falcao Salles J (2012) Fluctuations in ammonia oxidizing communities across agricultural soils are driven by soil structure and pH. Front Microbiol 3. doi:10.3389/fmicb.2012.00077
Di Giovanni GD, Watrud LS, Seidler RJ, Widmer F (1999) Comparison of parental and transgenic alfalfa rhizosphere bacterial communities using biolog GN metabolic fingerprinting and enterobacterial repetitive intergenic consensus sequence-PCR (ERIC-PCR). Microb Ecol 37(2):129–139. doi:10.1007/s002489900137
Grayston SJ, Wang S, Campbell CD, Edwards AC (1998) Selective influence of plant species on microbial diversity in the rhizosphere. Soil Biol Biochem 30(3):369–378. doi:10.1016/s0038-0717(97)00124-7
Latour X, Corberand T, Laguerre G, Allard F, Lemanceau P (1996) The composition of fluorescent pseudomonad populations associated with roots is influenced by plant and soil type. Appl Environ Microbiol 62(7):2449–2456
Ben-Hammouda M, Kremer RJ, Minor HC (1995) Phytotoxicity of extracts from sorghum plant components on wheat seedlings. Crop Sci 35(6):1652–1656
Czarnota MA, Paul RN, Dayan FE, Nimbal CI, Weston LA (2001) Mode of action, localization of production, chemical nature, and activity of sorgoleone: a potent PSII inhibitor in Sorghum spp. root exudates. Weed Technol 15(4):813–825
Weston LA, Harmon R, Mueller S (1989) Allelopathic potential of sorghum-sudangrass hybrid (sudex). J Chem Ecol 15(6):1855–1865
Coelho MRR, Marriel IE, Jenkins SN, Lanyon CV, Seldin L, O’Donnell AG (2009) Molecular detection and quantification of nifH gene sequences in the rhizosphere of sorghum (Sorghum bicolor) sown with two levels of nitrogen fertilizer. Appl Soil Ecol 42(1):48–53. doi:10.1016/j.apsoil.2009.01.010
Hai B, Diallo NH, Sall S, Haesler F, Schauss K, Bonzi M et al (2009) Quantification of key genes steering the microbial nitrogen cycle in the rhizosphere of sorghum cultivars in tropical agroecosystems. Appl Environ Microbiol 75(15):4993–5000. doi:10.1128/aem.02917-08
Coelho MRR, Da Mota FF, Carneiro NP, Marriel IE, Paiva E, Rosado AS et al (2007) Diversity of Paenibacillus spp. in the rhizosphere of four sorghum (Sorghum bicolor) cultivars sown with two contrasting levels of nitrogen fertilizer assessed by rpoB-based PCR-DGGE and sequencing analysis. J Microbiol Biotechnol 17(5):753–760
Mao Y, Yannarell AC, Mackie RI (2011) Changes in N-transforming Archaea and bacteria in soil during the establishment of bioenergy crops. PLoS One 6(9):12. doi:10.1371/journal.pone.0024750, e24750
Culman SW, DuPont ST, Glover JD, Buckley DH, Fick GW, Ferris H et al (2010) Long-term impacts of high-input annual cropping and unfertilized perennial grass production on soil properties and belowground food webs in Kansas, USA. Agr Ecosyst Environ 137(1–2):13–24, 10.1016/j.agee.2009.11.008
Liang C, EdC J, Duncan DS, Jackson RD, Tiedje JM, Balser TC (2012) Soil microbial communities under model biofuel cropping systems in southern Wisconsin, USA: impact of crop species and soil properties. Appl Soil Ecol 54:24–31. doi:10.1016/j.apsoil.2011.11.015
Arriola PE, Ellstrand NC (1996) Crop-to-weed gene flow in the genus Sorghum (Poaceae): spontaneous interspecific hybridization between johnsongrass, Sorghum halepense, and crop sorghum. S Bicolor American J Botany 83(9):1153–1159
Skinner K, Smith L, Rice PM (2000) Using noxious weed lists to prioritize targets for developing weed management strategies. Weed Sci 48(5):640–644
Warwick SI, Black LD (1983) The biology of Canadian weeds. 61. Sorghum halepense (L.) Pers. Canadian J Plant Sci 63(4):997–1014
Callaway RM, Thelen GC, Rodriguez A, Holben WE (2004) Soil biota and exotic plant invasion. Nat 427(6976):731–733
Reinhart KO, Callaway RM (2006) Soil biota and invasive plants. New Phytol 170(3):445–457. doi:10.1111/j.1469-8137.2006.01715.x
Acknowledgments
The authors thank their respective staff who participated in providing soil samples, photographs, GPS coordinates, and other background information for each of the crop and non-crop sampling locations. This research was funded in part by a United States Environmental Protection Agency (USEPA) Office of Research and Development National Health and Environmental Effects Research Laboratory intramural competitive award to LSW and RJF and by EPA contracts to Dynamac Corporation (EP-D-06-013 and EP-D-11–027). Mention of trade names or commercial products does not imply endorsement for use. The views of the authors do not necessarily reflect those of the Agency. This manuscript has undergone administrative and technical reviews to receive Agency approval for submission for publication in a peer-reviewed scientific journal.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Watrud, L.S., Reichman, J.R., Bollman, M.A. et al. Chemistry and Microbial Functional Diversity Differences in Biofuel Crop and Grassland Soils in Multiple Geographies. Bioenerg. Res. 6, 601–619 (2013). https://doi.org/10.1007/s12155-012-9279-y
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12155-012-9279-y