Abstract
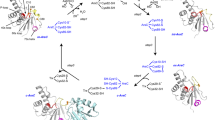
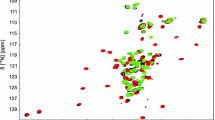
Peroxiredoxins (Prx) are ubiquitous enzymes that reduce peroxides as part of antioxidant defenses and redox signaling. While Prx catalytic activity and sensitivity to hyperoxidative inactivation depend on their dynamic properties, there are few examples where their dynamics has been characterized by NMR spectroscopy. Here, we provide a foundation for studies of the solution properties of peroxiredoxin Q from the plant pathogen Xanthomonas campestris (XcPrxQ) by assigning the observable 1HN, 15N, 13Cα, 13Cβ, and 13C′ chemical shifts for both the reduced (dithiol) and oxidized (disulfide) states. In the reduced state, most of the backbone amide resonances (149/152, 98 %) can be assigned in the XcPrxQ 1H–15N HSQC spectrum. In contrast, a remarkable 51 % (77) of these amide resonances are not visible in the 1H–15N HSQC spectrum of the disulfide state of the enzyme, indicating a substantial change in backbone dynamics associated with the formation of an intramolecular C48–C84 disulfide bond.


Similar content being viewed by others
References
Ådén J, Wallgren M, Storm P, Weise CF, Christiansen A, Schroder WP, Funk C, Wolf-Watz M (2011) Extraordinary μs–ms backbone dynamics in Arabidopsis thaliana peroxiredoxin Q. Biochem Biophys Acta 1814:1880–1890
Buchko GW, Daughdrill GW, de Lorimier R, Rao BK, Isern NG, Lingbeck JM, Taylor JS, Wold MS, Gochin M, Spicer LD, Lowry DF, Kennedy MA (1999) Interactions of human nucleotide excision repair protein XPA with DNA and RPA70ΔC327: chemical shift mapping and 15N NMR relaxation studies. Biochemistry 38:15116–15128
Leyns F, De Cleene M, Swings J, De Ley J (1984) The host range of the genus Xanthomonas. Bot Rev 50:305–355
Liao S-J, Yang C-Y, Chin K-H, Wang AHJ, Chou S-H (2009) Insights into the alkyl peroxide reduction pathway of Xanthomonas campestris bacterioferritin comigratory protein from the trapped intermediate–ligand complex structures. J Mol Biol 390:951–966
Maiti R, Van Domselaar GH, Zhang H, Wishart DS (2004) Superpose: a simple server for sophisticated structural superposition. Nucl Acids Res 32:W590–W594
Müller CW, Schlauderer GJ, Reinstein J, Schulz GE (1992) Adenylate kinase motions during catalysis: an energetic counterweight balancing substrate binding. Structure 4:147–156
Myler PJ, Stacy R, Stewart LJ, Staker BL, Van Voorhis WC, Varani G, Buchko GW (2009) The Seattle Structural Genomics Center for Infectious Disease (SSGCID). Infect Dis Drug Targets 9:493–506
Nelson KJ, Parsonage D, Hall A, Karplus PA, Poole LB (2008) Cysteine pKa values for the bacterial peroxiredoxin AhpC. Biochemistry 47:12860–12868
Perkins A, Poole LB, Karplus PA (2014) Tuning of peroxiredoxin catalysis for various physiological roles. Biochemistry 53:7693–7705
Rhee SG, Woo HA, Kil IS, Bae SH (2012) Peroxiredoxin functions as a peroxidase and a regulator and sensor of local peroxides. J Biol Chem 287:4403–4410
Sharma D, Rajarathnam K (2000) 13C NMR chemical shifts can predict disulfide bond formation. J Biomol NMR 18:165–171
Winterbourn CC (2008) Reconciling the chemistry and biology of reactive oxygen species. Nat Chem Biol 4:278–286
Yee A, Chang X, Pineda-Lucena A, Wu B, Semesi A, Le B, Ramelot T, Lee G, Bhattacharyya S, Gutierrez P, Denisov A, Lee C-H, Cort JR, Kozlov G, Liao J, Finak G, Chen L, Wishart D, Lee W, McIntosh LP, Gehring K, Kennedy MA, Edwards AM, Arrowsmith CH (2002) An NMR approach to structural proteomics. Proc Natl Acad Sci USA 99:1825–1930
Acknowledgments
This research was supported by the National Institute of Health R01 Grant number GM050389 (LBP and PAK). Part of this research was performed at the W.R. Wiley Environmental Molecular Sciences Laboratory (EMSL), a national scientific user facility located at Pacific Northwest National Laboratory (PNNL) and sponsored by US Department of Energy’s Office of Biological and Environmental Research (BER) program. Battelle operates PNNL for the US Department of Energy.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Buchko, G.W., Perkins, A., Parsonage, D. et al. Backbone chemical shift assignments for Xanthomonas campestris peroxiredoxin Q in the reduced and oxidized states: a dramatic change in backbone dynamics. Biomol NMR Assign 10, 57–61 (2016). https://doi.org/10.1007/s12104-015-9637-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12104-015-9637-8