Abstract
The C-terminus of the human adenosine A2A receptor differs from the other human adenosine receptors by its exceptional length and lack of a canonical cysteine residue. We have previously structurally characterized this C-terminal domain and its interaction with calmodulin. It was shown to be structurally disordered and flexible, and to bind calmodulin with high affinity in a calcium-dependent manner. Interaction with calmodulin takes place at the N-terminal end of the A2A C-terminal domain without major conformational changes in the latter. NMR was one of the biophysical methods used in the study. Here we present the HN, N, Cα, Cβ and C′ chemical shift assignments of the free form of the C-terminus residues 293–412, used in the NMR spectroscopic characterization of the domain.
Similar content being viewed by others
Biological context
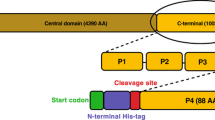
Human adenosine A2A receptor (A2AR) is a member of the large family of membrane-spanning G-protein-coupled receptors (GPCRs). GPCRs share a common topology with a transmembrane core composed of seven helices oriented parallel to the membrane. These helices are linked by three extra- and three intracellular loops. The C-terminus is intracellular. A2AR differs from other adenosine receptors by having a more than 80 residues longer C-terminus (A2A-ct). A2A-ct has 122 residues, whereas adenosine receptors A2B, A1 and A3 have 40, 38, and 34 residues, respectively. The A2A-ct amino acid sequence is highly conserved among species. Also, A2A-ct lacks the canonical cysteine residue at the end of the eighth helix. One or two cysteines are found at this position in the great majority of related proteins. This cysteine is a putative site for palmitoylation, which is known to promote membrane association. It has been suggested that the length of A2A-ct and the lack of membrane anchoring render the domain less constrained and more flexible, enabling A2A-ct-accessory protein interactions and A2AR subsequent signal transmission through G-protein-independent pathways (Keuerleber et al. 2011). Several proteins interacting with A2A-ct have been identified, including other signalling proteins such as ARNO/cytohesin-2 (Gsandtner et al. 2005), elementary in MAPK activation, NECAB2 (Canela et al. 2007), which enhances A2AR intracellular retention and signalling through MAPK, TRAX (Sun et al. 2006) involved in regulation of neurite growth, and calmodulin which modulates the function of A2AR-dopamine D2 receptor complexes (Woods et al. 2008).
We have recently shown that the C-terminal domain (A2A-ct, residues 293–412 of A2AR) is disordered and flexible (Piirainen et al. 2015), and can adopt different conformations depending on its surroundings and interaction partners. We also showed that calmodulin binds to the eighth helix of A2AR in a calcium-dependent manner. NMR spectroscopy was one of the biophysical methods used to characterize A2A-ct in free and calmodulin-bound forms. Here we report the backbone chemical shift assignments of the free form required for that study.
Methods and experiments
Production of uniformly 13C, 15N labelled A2AR C-terminal amino acids 293–412 has been described in Piirainen et al. (2015). All A2A-ct NMR spectra were measured in 95/5 % 50 mM HEPES, 250 mM NaCl, 1 mM TCEP-HCl, 5 mM CaCl2 pH 7.2/D2O buffer at 25 °C in a Shigemi micro cell on a Varian INOVA 800 MHz NMR spectrometer (Agilent, Santa Clara, California) equipped with a cryogenically cooled 1H, 13C, 15N probehead. Protein concentration was 0.6 mM. For the backbone assignment of A2A-ct we used both conventional HN-detected triple-resonance experiments 1H, 15N-HSQC, HNCACB, CBCA(CO)NH and HNCO (for Review see e.g., Permi and Annila 2004) as well as three-dimensional HN-detected experiments targeted to intrinsically disordered systems, i(HCA)CO(CA)NH (Mäntylahti et al. 2009) (HCA)CON(CAN)H and (HCA)N(CA)CO(N)H (Hellman et al. 2014). Spectra were processed with VnmrJ (Agilent, Santa Clara, California) and analyzed with Sparky (T. D. Goddard and D. G. Kneller, University of California, San Francisco).
Assignments and data deposition
For the assignment we employed a combination of common 3D HN-detected assignment spectra and 3D HN-detected experiments specially tailored for intrinsically disordered proteins (Mäntylahti et al. 2009; Hellman et al. 2014). Fast conformational averaging and chemical exchange and absence of structure-induced secondary chemical shifts result in a narrow HN chemical shift range as well as overlap of Cα and Cβ shifts, both commonly observed for IDPs. The tailored experiments exploit the chemical shift dispersion of the N and C′ directions and the favourable neighboring-residue effect on C′, and resolve ambiguities by separating intraresidual and sequential cross peaks to different spectra. Moreover, (HCA)CON(CAN)H and (HCA)N(CA)CO(N)H spectra enable linking the amino acid stretches flanking single proline residues that are highly abundant in intrinsically disordered proteins.
Omitting the C-terminal His-tag, an almost complete assignment of HN, N, C′, Cα and Cβ was reached with HNCACB, CBCA(CO)NH, HNCO and its intraresidual analogue i(HCA)CO(CA)NH supplemented with (HCA)CON(CAN)H and (HCA)N(CA)CO(N)H spectra. The former spectrum displays cross-peaks between Ni, C′i and H Ni and the latter between H Ni , C′i and Ni+1. Without assignments were left the first residue, Met292, the HN of the second residue, Arg293, the amide group of His306 and the carbon shifts of prolines 342 and 397. In the amino acid sequence these are the first of two consecutive prolines, GHPP and PEPP, the carbon chemical shift of which are not attainable by the used assignment spectra. Only the first, His416, and the last two histidines, His424 and His425, of the His-tag could be assigned. The 15Ni–13Ci plane of the (HCA)N(CA)CO(N)H spectrum of A2A-ct is shown in Fig. 1. Only few residues overlap in the 2D plane and all of those have distinct 1Hi+1 chemical shifts in the third dimension, enabling their unambiguous separation. The additional benefit from the (HCA)CON(CAN)H and (HCA)N(CA)CO(N)H spectra is that that the sequential walk is not interrupted by prolines of the amino acid sequence. In Fig. 2 is shown the assignment of residues Ser350–Ala–Pro–His–Pro–Glu of A2A-ct.
Assignment of A2A-ct shown in the (HCA)N(CA)CO(N)H spectrum. The Ni–Ci plane is displayed. Cross peaks of proline and glycine residues appear in opposite phase. Encircled cross peaks have no assignments and can originate from the His-tag, sample degradation or impurity. Cross-peaks of residues Arg304 and His338 are visible at a lower contour level. Six cross peaks are missing from the spectrum, namely those of His305, Lys390–Cys393 and Glu395
Sequential walk through prolines. Assignment of residues Ser350–Ala–Pro–His–Pro–Glu is shown with (HCA)CON(CAN)H and (HCA)N(CA)CO(N)H spectra at the bottom. In the latter spectrum cross peaks of Pro352 and Pro354 are aliased in the 15N dimension and thus of opposite phase. Their true 15N chemical shifts are given in parentheses. Further evidence for the reliability of the assignment is obtained from HNCO and i(HCA)CO(CA)NH shown on the top. Other assignments on the same plane are shown in grey and from succeeding planes in grey and italics
The 1H, 15N HSQC spectrum with backbone amide assignments as well as the secondary structure propensity of A2A-ct are shown in Piirainen et al. 2015. The chemical shifts have been deposited to the BioMagResBank database under accession number 25445.
References
Canela L, Luján R, Lluís C, Burgueño J, Mallol J, Canela EI, Franco R, Ciruela F (2007) The neuronal Ca2+-binding protein 2 (NECAB2) interacts with the adenosine A2A receptor and modulates the cell surface expression and function of the receptor. Mol Cell Neurosci 36:1–12
Gsandtner I, Charalambous C, Stefan E, Ogris E, Freissmuth M, Zezula J (2005) Heterotrimeric G protein-independent signaling of a G protein-coupled receptor. Direct binding of ARNO/cytohesin-2 to the carboxyl terminus of the A2A adenosine receptor is necessary for sustained activation of the ERK/MAP kinase pathway. J Biol Chem 280:31898–31905
Hellman M, Piirainen H, Jaakola V-P, Permi P (2014) Bridge over troubled proline: assignment of intrinsically disordered proteins using (HCA)CON(CAN)H and (HCA)N(CA)CO(N)H experiments concomitantly with HNCO and i(HCA)CO(CA)NH. J Biomol NMR 58:49–60
Keuerleber S, Gsandtner I, Freissmuth M (2011) From cradle to twilight: the carboxyl terminus directs the fate of the A2A-adenosine receptor. Biochim Biophys Acta 1808:1350–1357
Mäntylahti S, Tossavainen H, Hellman M, Permi P (2009) An intraresidual i(HCA)CO(CA)NH experiment for the assignment of main-chain resonances in 15N, 13C labeled proteins. J Biomol NMR 45:301–310
Permi P, Annila A (2004) Coherence transfer in proteins. Prog Nucl Magn Reson Spectr 44:97–137
Piirainen H, Hellman M, Tossavainen H, Permi P, Kursula P, Jaakola V-P (2015) Human adenosine A2A receptor binds calmodulin with high affinity in a calcium-dependent manner. Biophys J 108:903–917
Sun C-N, Cheng H-C, Chou J-I, Lee S-Y, Lin Y-W, Lai H-L, Chen H-M, Chern Y (2006) Rescue of p53 blockage by the A2A adenosine receptor via a novel interacting protein, translin-associated protein X. Mol Pharmacol 70:454–466
Woods AS, Marcellino D, Jackson SN, Franco R, Ferré S, Agnati LF, Fuxe K (2008) How calmodulin interacts with the adenosine A2A and the dopamine D2 receptors. J Proteome Res 7:3428–3434
Acknowledgments
This work was supported by the Academy of Finland Grant 259447 to PP.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Tossavainen, H., Hellman, M., Piirainen, H. et al. HN, N, Cα, Cβ and C′ assignments of the intrinsically disordered C-terminus of human adenosine A2A receptor. Biomol NMR Assign 9, 403–406 (2015). https://doi.org/10.1007/s12104-015-9618-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12104-015-9618-y