Abstract
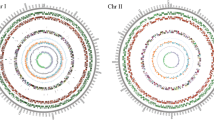
Whole-genome sequencing of bacterial pathogens has high impact on diagnosis and development of prophylaxis to human and veterinary disease. To study the genetic variations and relatedness among Brucella spp., we have examined the pan-genomic content of 18 Brucella complete genomes from NCBI using GView and Panseq server. Multiple sequence alignment of 16S rRNA gene sequences and multiple genome alignment of all 18 Brucella spp. showed high conservation at the nucleotide level. The core, accessory and unique gene content of Brucella spp. were estimated to be 2473, 7972 and 235 respectively. The presence of strain specific region in chromosome 1 of B. melitensis M5-90 and chromosome 2 of B. microti CCM 4915, B. pinnipedialis B2/94, and B. suis ATCC 23445 were identified. In addition, a stretch of genetic pool harboring 206 strain specific coding genes trans-located from chromosome 1 to chromosome 2 were also identified in the genome of B. suis ATCC 23445. Pan-genome analysis has also revealed the integration of strain specific 23 CDS (57.82 Kb) and 6 CDS (12 Kb) in B. pinnipedialis B2/94 and B. microti CCM 4915, respectively. Major pathogenic determinants like virB operon, non-classical lipopolysaccharide, two-component system BvrR/BvrS and cyclic β-1, 2-glucan synthase were unanimously found in all Brucella spp.. The virulence genes such as flagella, type III secretion system, hemolysins, outer membrane proteins, acvB, MViN and genes involved in eryrthritol metabolism were found in the accessory genome, which may guide the host specificity.







Similar content being viewed by others
References
Corbel MJ (1997) Brucellosis: an overview. Emerg Infect Dis 3:213–221
Czibener C, Ugalde JE (2012) Identification of a unique gene cluster of Brucella spp. that mediates adhesion to host cells. Microbes Infect 14:79–85
Whatmore AM (2009) Current understanding of the genetic diversity of Brucella, an expanding genus of zoonotic pathogens. Infect Genet Evol 9:1168–1184
DelVecchio VG, Kapatral V, Redkar RJ, Patra G, Mujer C, Los T, Ivanova N, Anderson I, Bhattacharyya A, Lykidis A, Reznik G, Jablonski L, Larsen N, D’Souza M, Bernal A, Mazur M, Goltsman E, Selkov E, Elzer PH, Hagius S, O’Callaghan D, Letesson JJ, Haselkorn R, Kyrpides N, Overbeek R (2002) The genome sequence of the facultative intracellular pathogen Brucella melitensis. Proc Natl Acad Sci USA 99:443–448
Chain PS, Comerci DJ, Tolmasky ME, Larimer FW, Malfatti SA, Vergez LM, Aguero F, Land ML, Ugalde RA, Garcia E (2005) Whole-genome analyses of speciation events in pathogenic Brucellae. Infect Immun 73:8353–8361
Franco AJ, Maurel D, Cotella O, Urrusuno JL (1999) Statistics on human brucellosis in the Republic of Argentina. Rev Argent Microbiol 31:52–55
Pappas G, Akritidis N, Bosilkovski M, Tsianos E (2005) Brucellosis. N Engl J Med 352:2325–2336
Lopez-Goni I, Garcia-Yoldi D, Marin CM, de Miguel MJ, Munoz PM, Blasco JM, Jacques I, Grayon M, Cloeckaert A, Ferreira AC, Cardoso R, Correa de Sa MI, Walravens K, Albert D, Garin-Bastuji B (2008) Evaluation of a multiplex PCR assay (Bruce-ladder) for molecular typing of all Brucella species, including the vaccine strains. J Clin Microbiol 46:3484–3487
Audic S, Lescot M, Claverie JM, Scholz HC (2009) Brucella microti: the genome sequence of an emerging pathogen. BMC Genom 10:352
Liu W, Fang L, Li M, Li S, Guo S, Luo R, Feng Z, Li B, Zhou Z, Shao G, Chen H, Xiao S (2012) Comparative genomics of Mycoplasma: analysis of conserved essential genes and diversity of the pan genome. PLoS One 7:35698
Medini D, Donati C, Tettelin H, Masignani V, Rappuoli R (2005) The microbial pan-genome. Curr Opin Genet Dev 15:589–594
Jacobsen A, Hendriksen RS, Aaresturp FM, Ussery DW, Friis C (2011) The Salmonella enterica pan-genome. Microb Ecol 62:487–504
Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, Ward NL, Angiuoli SV, Crabtree J, Jones AL, Durkin AS, Deboy RT, Davidsen TM, Mora M, Scarselli M, Ros IM, Peterson JD, Hauser CR, Sundaram JP, Nelson WC, Madupu R, Brinkac LM, Dodson RJ, Rosovitz MJ, Sullivan SA, Daugherty SC, Haft DH, Selengut J, Gwinn ML, Zhou L, Zafar N, Khouri H, Radune D, Dimitrov G, Watkins K, Connor KJ, Smith S, Utterback TR, White O, Rubens CE, Grandi G, Madoff LC, Kasper DL, Telford JL, Wessels MR, Rappuoli R, Fraser CM (2005) Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial pan-genome. Proc Natl Acad Sci USA 102:13950–13955
Mira A, Martin-Cuadrado AB, D’Auria G, Rodriguez-Valera F (2010) The bacterial pan-genome: a new paradigm in microbiology. Int Microbiol 13:45–57
Rodriguez-Valera F, Ussery DW (2012) Is the pan-genome also a pan-selectome? F1000Res. doi:10.3410/f1000research.1-16.v1
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and clustal X version 2.0. Bioinformatics 23:2947–2948
Nawrocki EP (2009) Structural RNA homology search and alignment using covariance models. Ph.D. thesis, Washington University in Saint Louis, School of Medicine
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Darling AE, Mau B, Perna NT (2010) progressiveMauve: multiple genome alignment with gene gain, loss, and rearrangement. PLoS One 5:11147
Xu Z, Hao B (2009) CVTree update: a newly designed phylogenetic study platform using composition vectors and whole genomes. Nucleic Acids Res 37:174–178
Page RD (1996) TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12:357–358
Petkau A, Stuart-Edwards M, Stothard P, Van Domselaar G (2010) Interactive microbial genome visualization with GView. Bioinformatics 26:3125–3126
Laing C, Buchanan C, Taboada EN, Zhang Y, Kropinski A, Villegas A, Thomas EJ, Gannon PJ (2010) Pan-genome sequence analysis using Panseq: an online tool for the rapid analysis of core and accessory genomic regions. BMC Bioinformatics 11:461
Bohlin J, Snipen L, Cloeckaert A, Lagesen K, Ussery D, Kristoffersen AB, Godfroid J (2010) Genomic comparisons of Brucella spp. and closely related bacteria using base compositional and proteome based methods. BMC Evol Biol 10:249
Halling SM, Peterson-Burch BD, Bricker BJ, Zuerner RL, Qing Z, Li LL, Kapur V, Alt DP, Olsen SC (2005) Completion of the genome sequence of Brucella abortus and comparison to the highly similar genomes of Brucella melitensis and Brucella suis. J Bacteriol 187:2715–2726
Banai M, Corbel M (2010) Taxonomy of Brucella. Open Vet Sci J 4:85–101
Lapaque N, Moriyon I, Moreno E, Gorvel JP (2005) Brucella lipopolysaccharide acts as a virulence factor. Curr Opin Microbiol 8:60–66
Wattam AR, Foster JT, Mane SP, Beckstrom-Sternberg SM, Beckstrom-Sternberg JM, Dickerman AW, Keim P, Pearson T, Shukla M, Ward DV, Williams KP, Sobral BW, Tsolis RM, Whatmore AM, O’Callaghan D (2014) Comparative phylogenomics and evolution of the brucellae reveal a path to virulence. J Bacteriol 196:920–930
Cardoso P, Macedo G, Azevedo V, Oliveira S (2006) Brucella spp noncanonical LPS: structure, biosynthesis, and interaction with host immune system. Microb Cell Fact 5:13
Detilleux PG, Deyoe BL, Cheville NF (1990) Penetration and intracellular growth of Brucella abortus in non phagocytic cells in vitro. Infect Immun 58:2320–2328
Ko J, Splitter GA (2003) Molecular host-pathogen interaction in brucellosis: current understanding and future approaches to vaccine development for mice and humans. Clin Microbiol Rev 16:65–78
Farlow J, Filippov AA, Sergueev KV, Hang J, Kotorashvili A, Nikolich MP (2014) Comparative whole genome analysis of six diagnostic Brucellaphages. Gene 541:115–122
Hammerl JA, Al Dahouk S, Nockler K, Gollner C, Appel B, Hertwig S (2014) F1 and tbilisi are closely related Brucellaphages exhibiting some distinct nucleotide variations which determine the host specificity. Genome Announc 2:01250-13
Sola-Landa A, Pizarro-Cerda J, Grillo MJ, Moreno E, Moriyon I, Blasco JM, Gorvel JP, López-Goni I (1998) A two-component regulatory system playing a critical role in plant pathogens and endosymbionts is present in Brucella abortus and controls cell invasion and virulence. Mol Microbiol 29:125–138
Guzman-Verri C, Manterola L, Sola-Landa A, Parra A, Cloeckaert A, Garin J, Gorvel JP, Moriyon I, Moreno E, Lopez-Goni I (2002) The two-component system BvrR/BvrS essential for Brucella abortus virulence regulates the expression of outer membrane proteins with counterparts in members of the Rhizobiaceae. Proc Natl Acad Sci USA 99:12375–12380
Lopez-Goni I, Guzman-Verri C, Manterola L, Sola-Landa A, Moriyon I, Moreno E (2002) Regulation of Brucella virulence by the two-component system BvrR/BvrS. Vet Microbiol 90:329–339
Manterola L, Moriyon I, Moreno E, Sola-Landa A, Weiss DS, Koch MH, Howe J, Brandenburg K, Lopez-Goni I (2005) The lipopolysaccharide of Brucella abortus BvrS/BvrR mutants contains lipid A modifications and has higher affinity for bactericidal cationic peptides. J Bacteriol 187:5631–5639
Arellano-Reynoso B, Lapaque N, Salcedo S, Briones G, Ciocchini AE, Ugalde R, Moreno E, Moriyon I, Gorvel JP (2005) Cyclic β-1,2-glucan is a Brucella virulence factor required for intracellular survival. Nat Immunol 6:618–625
Guidolin LS, Ciocchini AE, Iannino IN, Ugalde RA (2009) Functional mapping of Brucella abortus cyclic beta-1,2-glucan synthase: identification of the protein domain required for cyclization. J Bacteriol 191:1230–1238
Verstreate DR, Creasy MT, Caveney NT, Baldwin CL, Blab MW, Winter AJ (1982) Outer membrane proteins of Brucella abortus: isolation and characterization. Infect Immun 35:979–989
Smith H, Williams AE, Pearce JH, Keppie J, Harris-Smith PW, Fitz-George RB, Witt K (1962) Foetal erythritol: a cause of the localization of Brucella abortus in bovine contagious abortion. Nature 193:47–49
Web Resources
NCBI: ftp://ncbi.nlm.nih.gov/
SSU-ALIGN: http://selab.janelia.org/software/ssu-align/
Treeview software: http://taxonomy.zoology.gla.ac.uk/rod/treeview.html
GView server: https://server.gview.ca
Panseq server: http://lfz.corefacility.ca/panseq/
Acknowledgments
The work was financially supported by the Department of Biotechnology, Govt. of India under DBT-Network Project on Brucellosis. One of the authors JS thank DBT for financial support. Authors also acknowledge the UGC-CAS, UGC-CEGS, UGC-NRCBS, DBT-IPLS and DST-PURSE programs of SBS, MKU.
Conflict of interest
The authors have declared that no conflict of interests exists.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
12088_2014_486_MOESM1_ESM.xlsx
The core genes, accessory genes and the unique genes were denoted with different colour fonts (Core genes- red, Accessory genes-green and Unique genes-violet). Supplementary material 1 (XLSX 645 kb)
Rights and permissions
About this article
Cite this article
Sankarasubramanian, J., Vishnu, U.S., Sridhar, J. et al. Pan-Genome of Brucella Species. Indian J Microbiol 55, 88–101 (2015). https://doi.org/10.1007/s12088-014-0486-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12088-014-0486-4