Abstract
Evolutionary trees are slowly but surely making their way into high school classrooms, and teachers are increasingly gaining access to resources that may help in fostering a better understanding of how to interpret or read phylogenies. Nevertheless, instructional material on the basic principles behind building evolutionary trees is still sorely lacking. Here we provide a brief overview of the highly classroom compatible tree building method known as maximum parsimony as well as a novel lab.
Similar content being viewed by others
“Community of descent is the hidden bond which naturalists have been unconsciously seeking, and not some unknown plan of creation, or the enunciation of general propositions and the mere putting together and separating of objects more or less alike” (Darwin 1872)
Evolutionary treesFootnote 1 are becoming increasingly popular in both scientific publications and educational resources. Nevertheless, there is a great deal of confusion with regards to how a tree is built and the rationale behind one of the more practical tree-building or “phylogenetic” methods, maximum parsimony, even amongst scientists! Although the ability to properly read and interpret evolutionary trees can be considered one of the cornerstones of biological literacy, understanding how they are constructed is also important because an evolutionary hypothesis is only as strong as the data supporting it and not even the allure of genetic precision can transform junk into treasure.
With this in mind, The Gummy Tree Challenge was created to demonstrate the fundamental principles of the phylogenetic method using resources that are accessible to all teachers, while offering the novelty of a fun, interactive activity for students. Recommended gummy candies to use for this exercise, as well as an answer key and student worksheet, are attached at the end of this document. To control the outcome, characters were selected beforehand, which is obviously not what would occur in a real study but makes it practical for teaching purposes. As a result, if you do not use the same candies, you will need to slightly adjust your characters. It is suggested, however, to select treats that demonstrate a gradual transition in physical properties (i.e., diversity in gummy bears with regards to color, texture, size, etc.). Also, it is probably best if re-designing aspects of this lab to limit your “conflicting” characters to just one in order to ensure maximum likelihood of student understanding. As a final note, I often have the students do this in groups (two to five) and issue a challenge for the most strongly supported hypothesis or “best tree” with a prize of more gummy bears! The remainder of this article will now serve to explain the phylogenetic method.
So how do we come up with all these evolutionary trees? Is it purely intuition and experience or is there a scientific methodology behind it? Contrary to popular belief, we do not rely on “overall similarity” to recover the “hidden bond” between organisms.Footnote 2 Why? Because there are different ways to be similar and some similarities can obscure the actual evolutionary relationships between species within this “community of descent.” We call a similarity that is not due to recent common ancestry a “homoplasy” or analogy instead of a homology or homologous character (i.e., the arm bones shared by all tetrapods are homologous). Homoplasies are the products of convergent or parallel evolution. Some prominent examples include: homeothermy or “warm-bloodedness” (mammals and birds), winged flight (i.e., insects, birds, bats, pterosaurs, etc.), multicellularity (i.e., bacteria, plants, animals, fungi), fusiform bodies (i.e., fish, ichthyosaurs, whales, manatees, etc.), bilaterally symmetrical flowers (orchids and lupines), limblessness in squamates (snakes + lizards), sabreteeth (marsupials and placentals), “fossorial” digging appendages, webbed feet, and so on. But how do we distinguish homology from homoplasy? Better yet, how do we avoid circular reasoning (i.e., using homology to determine the topology of the tree and then using the tree to determine subsequent homology)?
There are four ways to be similar. The first three refer to varying levels of relative homology: (1) shared, general homologies or symplesiomorphies (i.e., the vertebral column shared between salmon, humans, and gorillas are homologous but will not help determine that humans and gorillas are more related to one another than either is to salmon); (2) shared, special homologies (i.e., the hair and mammary glands shared by all living mammals); (3) shared, unique homologies or autapomorphies (i.e., features unique to members of a species that help in diagnostic identification). Only special homologies or synapomorphies will help resolve the interrelationships between species. Using overall similarity often causes organisms with less derived features to be artificially lumped together based on general similarities (i.e., all single-celled eukaryotes placed in the artificial group “Protista” even though some of them are more related to multicellular plants, animals, or fungi, respectively). The fourth way to be similar is through convergent or parallel evolution, which results in homoplasy, as was discussed earlier. So how do we distinguish general from special homology and homology from homoplasy?
Before you can get started, you need to establish the taxa (= any named group of organisms) to be included in your study (the “ingroup”). These are the organisms whose genealogical relationships are of interest to you as a researcher (a minimum of three taxa is required). Once you have your ingroup, you will need an “outgroup” to distinguish general from special homologous traits. If the state of a character exists outside the ingroup, it is a general trait and will not help resolve relationships within the ingroup. The “sister group” is the most closely related outgroup and usually multiple outgroups are used to establish the ancestral state of a given character with respect to the ingroup. For example, if we were studying mammals in general, reptiles and amphibians would make suitable outgroups for comparing character traits. The fossil record confirms that amphibians have been in existence longer than mammals, while other studies have independently confirmed that reptiles represent our sister group and are therefore more likely to share the most similar properties with mammals (that is, when comparing only living organisms). This is non-circular because homology status (general vs. special) is being assigned based on the properties of organisms whose relationships are not being assessed. Yes, in case you were wondering, the outgroup to all life is indeed a rock!Footnote 3 In the Gummy Tree Challenge, the burrowing smooth worm (Wormus smoothus) represents the outgroup to an ingroup of four gummies with unresolved relationships. Now that we have our ingroup and outgroup, we can begin:
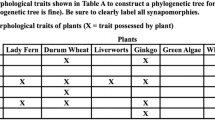
The first step is to presume a priori that all similarities within the ingroup represent homologies (i.e., a hypothesis of homology). This is the equivalent of a “null hypothesis” that we expect to be falsified if evolution has been complex. Step two is to use outgroup comparison to distinguish general from special homologous traits. Essentially, if a given character is found in both the ingroup and outgroup(s), it is designated as being in an ancestral state or plesiomorphic condition and a zero (“0”) is placed in our simple example of a binary “transformation” matrix. If a trait is new to the ingroup, it is designated as being in a derived or changed state (apomorphic) and assigned a “1.” In doing so, independent character evolution statements are made that will hopefully settle on an unequivocal or unambiguous pattern of genealogical descent.Footnote 4 Whenever more than one ingroup taxa shares a derived character, this represents a potential special homology (a possible synapomorphy). Step three is to group taxa according to all possible synapomorphies. One recommended strategy is to literally circle all the 1's for a given trait and group the taxa accordingly character by character (i.e., Bearus bigus, Bearus roughus, and Bearus redus are all bear-shaped; B. roughus and B. redus are both small and red). Step four is to combine the patterns of relatedness across all characters. If you encounter a relationship conflict (i.e., B. roughus and Dinaris sievus grouped by “rough texture” versus B. roughus and B. redus grouped by “bear-shape,” “small size,” and “redness”), choose the evolutionary relationship supported by the largest number of traits and thus the fewest number of proposed changes.
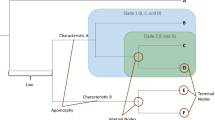
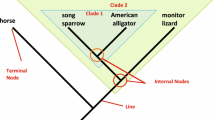
In the case of competing hypotheses (which is what every tree is), the tree with the fewest number of proposed changes for a given data set is judged to be the best. This is called the Principle of Parsimony or Ockham’s Razor, which states that, “entities should not be multiplied beyond necessity” or as often used in science, “if you have two competing theories, preference should be given to the simpler one until more evidence comes along” (i.e., the tree with five evolutionary steps proposes fewer changes than the tree of sevenFootnote 5). The fifth and final step is to interpret inconsistent results as falsifications of our original homology hypothesis. Therefore, each character that is in conflict with the consensus pattern is designated as a homoplasy (i.e., rough texture) after the tree is already built (Fig. 1). This is non-circular because “homologies, which indicate phylogenetic relationships, are determined without reference to a phylogeny, while homoplasies, which are inconsistent with phylogeny, are determined as such by reference to the phylogeny” (see Brooks and McLennan 2002 and references therein for more detail).
“Homologous parts tend to vary in the same manner, and homologous parts tend to cohere” (Darwin 1872)
Of course this does NOT mean that the evolution of life has been simplistic or parsimonious. In fact, we think the history of life has been so complex that our original null hypothesis of homology will be falsified to reveal conflicts in the data set and show us just how non-parsimonious evolution has actually been. If evolution has occurred in the simplest manner possible, there would never be any conflicting characters and overall similarity would perfectly correlate with a single, unambiguous answer or, in this case, tree. The principle of parsimony is only invoked afterwards to decide between competing evolutionary hypotheses and is justified on the basis that DNA replication rates are much higher than mutation rates. Therefore, we expect homologies to “cohere” or become inherited together as part of a single pattern, while homoplasies, far fewer in number, will be revealed to us as outliers. In a nutshell, the conservative, replicative nature of living things ensures that organisms will be more similar than they are different. This also means that the history of life’s origin and diversification will be preserved in the characters observed in species, living and fossil (Brooks and McLennan 2002), and by evaluating these signals, we are able to reveal the relationships of the past. While the resulting classification of living things based on evolutionary trees may be new and unfamiliar to most, it is now founded on a practice that lends itself to testing, falsification, and repetition, the very basis of good science.
Notes
Synonymous with phylogenies or cladograms
That’s a debunked “evolution-free” method called phenetics and often employs such techniques as DNA–DNA hybridization and neighbor-joining.
Minerals such as mackinawite ((Fe,Ni)xS) and greigite (NiS2[Fe4S4]S2Fe) are capable of catalyzing reactions that are eerily similar to the simplest carbon-fixing metabolic reactions performed by certain groups of bacteria, releasing “free energy” to perform additional work in the process. The enzymes used in the reductive acetyl–CoA pathway (hydrogen (H2) + carbon dioxide (CO2) → acetyl–CoA (CH3CO∼ScoA) + water (H2O)) by these “acetogens” have iron–sulfur cores that appear homologous to the structure of these minerals!
Note: plesiomorphic and apomorphic are terms used when making individual character evolution arguments. We cannot determine homology status (i.e., symplesiomorphy, synapomorphy, autapomorphy) until looking at the entire data set. For example, if we were looking at the character “body hair coverage,” we could say that “reduced body hair” is a derived or “apomorphic” condition of human primates, and after analyzing a larger data set, come to the conclusion that reduced body hair is an “autapomorphy” of human primates.
Otherwise, bear shape, small size, and redness would have to evolve independently in B. roughus.
References
Brooks DR, McLennan DA. The nature of diversity: an evolutionary voyage of discovery. 1st ed. Chicago: The University of Chicago Press; 2002.
Darwin C. The origin of species. 6th ed. London: John Murray; 1872.
Author information
Authors and Affiliations
Corresponding author
Appendix
Appendix


Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License ( https://creativecommons.org/licenses/by-nc/2.0 ), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Kumala, M. The Gummy Tree Challenge—Building Connections One Treat at a Time. Evo Edu Outreach 3, 520–525 (2010). https://doi.org/10.1007/s12052-010-0275-4
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12052-010-0275-4