Abstract
The line of descent that includes all living mammals extends back in time over 300 million years. Many of the ancient relatives of mammals that fall along this line are very different in appearance from living mammals and are frequently mistaken for reptiles such as dinosaurs. This misconception is reinforced by the fact that these animals are often referred to as “mammal-like reptiles,” a term reflecting outdated methods for classifying organisms. In reality, these ancient mammal-relatives, known as synapsids, are more closely related to living mammals than they are to any reptiles. Evolutionary trees, which depict patterns of descent from common ancestors among organisms, are very useful for understanding why this is the case and for reconstructing the evolutionary histories of many of the unique characters found in mammals. Here, I provide an introduction to evolutionary trees and their implications for understanding the relationships between mammals, synapsids, and reptiles. This is followed by a review of synapsid diversity and a discussion of how evolutionary trees can be used to investigate when in synapsid history different mammalian characteristics first appeared.
Similar content being viewed by others
Introduction
The sail-backed carnivore Dimetrodon (Fig. 1) frequently can be found on the pages of dinosaur books and lurking among the plastic dinosaurs for sale at museum gift shops. Nevertheless, Dimetrodon is not a dinosaur; it became extinct about 60 million years before the first dinosaurs evolved (almost the same amount of time that separates humans from Tyrannosaurus rex), and it is more closely related to living mammals, including humans, than it is to any extinct or living reptile. The idea that Dimetrodon is not a dinosaur but a mammal relative instead, is familiar to idea to vertebrate paleontologists: It was first formulated in the second half of the nineteenth century (Cope 1878) and is well supported by diverse types of evidence. Dimetrodon belongs to the array of fossil vertebrates that document in stunning detail the origin of mammals and the evolution of many mammalian characteristics such as a single lower jaw bone, complex teeth for chewing, a novel jaw muscle anatomy, large brains, and three tiny middle ear bones. Yet, many people, ranging from college students to my friends and family to visitors at The Field Museum are surprised to learn that Dimetrodon is not a dinosaur but part of our family tree. Dinosaur Dimetrodon is a persistent and widespread misconception. No wonder a T-shirt sold at the annual meeting of the Society of Vertebrate Paleontology features an angry Dimetrodon shouting, “I am not a dinosaur!”
Dimetrodon is a member of the large group of terrestrial vertebrates or tetrapods known as the Synapsida. Synapsids include all living mammals as well as a diverse range of extinct relatives extending back to the latter parts of the Carboniferous Period of Earth history, about 305 million years ago. They are justifiably well known as one of the great evolutionary sequences preserved in the fossil record, illustrating the evolution of mammals from an ancient, somewhat lizard-like ancestor. The nature of this transition has been reviewed by a number of authors at differing levels of complexity and detail (e.g., Hopson 1987, 1991, 1994, 2001; Hotton 1991; Rubidge and Sidor 2001; Kielan-Jaworowska et al. 2003; Martin 2004; Benton 2005; Kemp 2005; Prothero 2007), and my goal is not to simply repeat the material available in those works. Instead, I will discuss two perhaps more fundamental issues that I believe often lead to confusion about the synapsid fossil record and how it informs paleontologists about the evolution of mammals. In both cases, evolutionary trees, which show how different organisms are related to one another, are key for organizing information and gaining deeper understanding.
The first issue concerns the relationship between mammals and other, so-called non-mammalian synapsids, as well as the relationship between Synapsida as a whole and other groups of tetrapods. Popular treatments often are not clear on the fact that non-mammalian synapsids differ from reptiles and why this is the case. Imprecise and out-dated language which does not reflect current scientific thinking, such as “mammal-like reptile” or “reptile-to-mammal transition”, also frequently creeps into the discussion. When combined with the fact that many early synapsids, including Dimetrodon, superficially appear more reptilian than mammalian, these problems obscure the true nature of some synapsids and their evolutionary significance. Therefore, I will endeavor to clarify where synapsids like Dimetrodon fall on the great family tree of life, how we know they belong there, and why their position means that they are not reptiles or dinosaurs.
The second topic I will address is synapsid diversity and how it relates to the evolution of mammals. Because the synapsid fossil record provides such a detailed view of the evolution of mammals, synapsid diversity is often portrayed as an almost linear sequence, with each member of the sequence possessing a greater number of mammal-like features than the last. However, such linear sequences are rarely accurate (see MacFadden 1992 and Prothero 2007 for discussions of “classic” examples of linear evolution in horses and hominids that have been shown to be inaccurate), and this is true for synapsids. Synapsids existed for over 80 million years before the first mammals evolved, and during this time, non-mammalian synapsids evolved a fascinating array of shapes, sizes, and ways of life. These forms include large sabre-toothed carnivores, herbivores with turtle-like beaks, carnivores and herbivores with tall sails on their backs, specialized burrowers, small weasel-like carnivores, hippo-sized herbivores with thickened skulls that may have been used for head-butting, and even a carnivorous species that might have been venomous. This diversity is sometimes overlooked because much of it is peripheral to the evolution of mammals. At the same time, its existence can lead to questions and confusion. For example, if Dimetrodon is our relative, and it had a sail on its back, does that mean our direct ancestors did too? To answer this question, I will describe how evolutionary trees can be used to infer where in synapsid history particular mammalian characters evolved, as well as to recognize when a feature of interest represents an evolutionary innovation peculiar to a specific subgroup of synapsids, and therefore is not of direct relevance to the evolution of mammals.
Evolutionary Trees and Synapsids or Why Mammal-like Reptile is a Misnomer
The Basics of Evolutionary Trees
Dimetrodon and other non-mammalian synapsids often are referred to as mammal-like reptiles, and the evolution of mammals from an earlier synapsid ancestor is sometimes described as the reptile-to-mammal transition. These phrases are misleading because they imply that non-mammalian synapsids are somehow akin to living reptiles, such as lizards, crocodiles, or snakes, and that reptiles are ancestors of mammals. Both of these ideas are incorrect, and the easiest way to see why this is the case is to consult an evolutionary tree or phylogeny.
Evolutionary trees have existed as long as the science of evolutionary biology itself. Famously, the only figure in Darwin’s On the Origin of Species (1859) is an evolutionary tree. In the last four decades, the use of phylogenies as a framework for testing hypotheses and answering questions (so-called tree thinking) has revolutionized many areas of biology and paleontology, and it is now a central part of the biological sciences (O’Hara 1988, 1997; for a historical treatment of some of the debates surrounding the rise of tree thinking, see Hull 1988). Accompanying this revolution has been a fundamental shift in how scientists classify organisms, and this in turn has rendered terms such as mammal-like reptile obsolete.
At their most basic, evolutionary trees are diagrams that depict how recently three or more organisms shared a common ancestor (a useful introduction to phylogenies and tree thinking can be found in Gregory 2008). For example, in the simple phylogeny depicted in Fig. 2, we can see that humans and cats share a common ancestor because the branches of the tree leading up to them connect at their base (node 5). We also can see that crocodiles and turtles share a common ancestor because their branches connect at their base (node 4). Note, however, that the connection point (or node) between the branches leading up to turtles and crocodiles on the one hand, and cats and humans on the other, lies farther down the tree, at node 3. This implies that cats, humans, turtles, and crocodiles all share a common ancestor (node 3), but that this ancestor is older than the common ancestor of turtles and crocodiles (node 4) or the common ancestor of cats and humans (node 5). In other words, the phylogeny tells us that cats and humans share a more recent common ancestor with each other than either does with crocodiles or turtles, and that because of this more recent common ancestor, cats and humans are more closely related to each other than either is to turtles or crocodiles. Using the same logic, we can see that cats, humans, turtles, and crocodiles all share a more recent common ancestor (node 3) with each other than any does with salamanders and that salamanders, crocodiles, turtles, humans, and cats share a more recent common ancestor (node 2) with each other than any does with lungfish. Note that in all of this discussion, the relationships of particular organisms are always referred to relative to at least one other organism. This is because phylogenetic relationships are always relative. In other words, if we have only two organisms or groups, we can only state that they share a common ancestor, but we cannot state whether that common ancestor is recent or distant. Once we add in at least one more organism or group, however, we have a reference point that allows us to state that two of the organisms share a more recent common ancestor with each other (i.e., they are more closely related to each other) than either does with the third.
An evolutionary tree or phylogeny depicting the patterns of shared common ancestry between lungfish, salamanders, crocodiles, turtles, cats, and humans. The connection points between branches or nodes represent the most recent common ancestors of the animals found on the branches they connect. See text for details
An important question to ask at this point is how do we reconstruct the patterns of descent from common ancestors that are represented by an evolutionary tree? After all, we cannot see the relationships between different organisms directly. The answer is that scientists infer patterns of relationship based on the distribution of characters among a set of organisms of interest. Characters used in this process can take many forms, including skeletal features, aspects of soft tissue anatomy at both the microscopic and macroscopic levels, and DNA sequences. Typically, a large amount of character data will be collected, and then a phylogeny will be sought that best explains the evolution of the greatest number of characters given a specific optimality criterion, such as minimizing the number of hypothesized evolutionary changes or the best fit to an independently derived model of how DNA sequences change over time (technical information on how phylogenies are constructed can be found in Kitching et al. 1998; Felsenstein 2004, and Huelsenbeck and Ronquist 2005). Special attention is usually given to characters that all members of a particular group inherited from their most recent common ancestor because those characters are particularly useful for recognizing if newly discovered organisms are members of that group. So, in a sense, the characters organisms possess are something like tags indicating who their ancestors were, and scientists increase the likelihood that they have reconstructed an accurate evolutionary tree by finding the tree that best explains the evolution of the greatest number of characters.
The spread of tree thinking has had a profound effect on taxonomy and systematics, the sciences concerned with describing and naming species, and the placing of those species into larger named groups of organisms. Specifically, scientists now almost always try to recognize and name groups of species that are based only on patterns of common ancestry discovered through the construction of phylogenies, whereas older methods frequently conflated patterns of ancestry, the presence or absence of supposed “key” characters, and qualitative notions of how advanced different species were [the PhyloCode (Cantino and de Queiroz 2007), a proposed new set of rules for constructing names for organisms, represents one of the most fully developed results of this shift]. For example, Kielan-Jaworowska et al. (2003) recently defined mammals as “[the group] defined by the shared common ancestor of Sinoconodon, morganucodontans, Monotremata, Marupialia, and Placentalia, plus any extinct taxa shown to be nested with this [group]...” (p.2), a definition based on patterns of shared ancestry that are shown in a phylogeny (Fig. 3). In contrast, older definitions such as that of Simpson (1960) focused on the possession of particular characters, such as a single jaw bone or the presence of three middle ear bones, with less concern for whether the animals grouped together under such a definition included all descendants of a recent common ancestor. This methodological change is particularly important in the case of non-mammalian synapsids. Under older classification schemes, these animals were grouped with the reptiles because they lacked supposedly key mammalian characters, despite the fact that they are more closely related to mammals than to any reptiles. This is why non-mammalian synapsids often appear in older works describing the morphology, origins, and relationships of reptiles (e.g., Romer 1956; Carroll 1969a, b, 1970).
Simplified phylogeny of synapsids demonstrating how the definition provided by Kielan-Jaworowska et al. (2003; the common ancestor of Sinoconodon, morganucodontans, monotremes, marsupials, and placentals, plus any species nested within this group) delimits mammals. The definition uses patterns of common ancestry found in the phylogeny to denote which animals belong to the group Mammalia and which animals fall outside of that group. Names shown in bold are those used in the definition of Mammalia
Where Do Synapsids Fit?
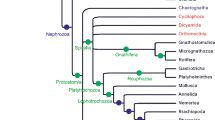
If we consider a phylogeny of living tetrapods and their close relatives (Fig. 4a), we can see that tetrapods are comprised of at least three and possibly four main groups, depending on whether caecilians, worm-like tetrapods found in the tropics, are more closely related to frogs and salamanders, as traditionally thought, or if they share a more recent common ancestor with mammals and reptiles (e.g., compare the trees of Ruta and Coates 2007 to that of Anderson et al. 2008). This uncertainty notwithstanding, the tree makes clear the relationships of living synapsids and reptiles: The three living synapsid groups, montreme mammals (platypus and echidnas), marsupial mammals (kangaroos, koalas, possums, and their relatives), and placental mammals (dogs, bats, whales, horses, humans, and their relatives) all are more closely related to each other than they are to any reptiles (i.e., they share a more recent common ancestor with each other than any living reptiles). In other words, reptiles and synapsids (represented among living animals by mammals) represent two distinct lines of descent from a common ancestor, and neither group is directly ancestral to the other. To claim that reptiles are ancestral to mammals would be similar to claiming that your cousin is ancestral to you. Both you and your cousin share common ancestors, your grandparents, but separate lines of descent lead from them to you and your cousin, one passing through your aunt or uncle, the other passing through your father or mother.
a Simplified phylogeny of living terrestrial vertebrates (or tetrapods). Note that the three living mammal groups (monotreme mammals, marsupial mammals, and placental mammals) all share a more recent common ancestor with each other than any does with the reptiles and that mammals and reptiles represent separate lines of descent from a common ancestor. Also note how patterns of shared ancestry are used to delimit the groups Mammalia, Reptilia, Amniota, and Tetrapoda. The position of caecilians on this tree is uncertain; they may belong on the branch with frogs and salamanders or they may belong on the branch leading to the Amniota [e.g., compare the phylogenies of Ruta and Coates (2007) to that of Anderson et al. (2008)]. b Simplified phylogeny of tetrapods to which a number of extinct amniotes have been added. Groups with living members are shown in bold. Note that birds are most closely related to non-avian saurischian dinosaurs on this tree, reflecting that birds are descendants of a dinosaur ancestor. Also note the large number of extinct groups of non-mammalian synapsids, which are descendants of the common ancestor of all synapsids, and thus are more closely related to living mammals than they are to any reptiles. In turn, this fact means that terms like mammal-like reptile do not accurately describe these animals because they are not part of the reptile line of descent. Dimetrodon is a sphenacodontid
Now, let us add some fossils to our evolutionary tree (Fig. 4b). A noteworthy change in the reptile portion of the tree that takes place when we do this is that living birds now share a common ancestor with non-avian saurishcian dinosaurs. This is because birds are descendants of a group of saurischian (“lizard-hipped”) dinosaurs called theropods, making them dinosaurs themselves and allowing us to draw a distinction between non-avian dinosaurs (i.e., all those dinosaurs that are not birds) and “avian dinosaurs,” birds themselves (an excellent review of the dinosaurian origin of birds can be found in Chiappe 2007). More important in the context of the current paper are the many groups of extinct synapsids and early mammals known from the fossil record that are added to the branch leading up to extant mammals. All of the groups on this branch below the node labeled “Mammalia” are non-mammalian synapsids. That is, they are descendants of the most recent common ancestor of all synapsids, but not the most recent common ancestor of all mammals.
In the past, non-mammalian synapsids were often colloquially referred to as mammal-like reptiles. They were “mammal-like” because paleontologists understood that they were related to mammals and provided insight into the latter group’s evolution, but they were “reptiles” because they lacked key characters that defined mammals, such as a single jaw bone or three middle ear bones. Examining the phylogeny in Fig. 4b shows why this terminology has been abandoned by scientists as tree thinking has become common and taxonomic groups have come to be defined by patterns of shared ancestry. Non-mammalian synapsids are descended from the most recent common ancestor of all synapsids, and not the most recent common ancestor of reptiles, making them by definition part of a line of descent that is separate from all reptiles. The fact that some of the earliest synapsids, such as Dimetrodon (which is a member of the synapsid subgroup called sphenacodontids in Fig. 4b), superficially resemble living reptiles in some respects does not overturn this underlying pattern of common ancestry.
Tree thinking also helps to clarify the nature of the evolution of mammals and many of their distinctive characters. Using outdated taxonomic concepts and terminology such as “mammal-like reptile” confuse the issue because they suggest that reptiles are ancestors of mammals. In turn, this can lead to questions such as, “if reptiles are ancestors of mammals, why are reptiles still alive today?” and “where are the missing links between reptiles and mammals?” Once again, the answers to these questions are obvious if we consult our evolutionary tree (Fig. 4b). Reptiles are not ancestors of mammals; they are part of a separate line of descent from a common ancestor, so their existence in the modern world is no more surprising than the fact that you and your cousin both exist today. There are no “missing links” between reptiles and mammals for exactly the same reason. Because reptiles and synapsids (including mammals) are two separate lines of descent, the link between the groups is the common ancestor they share (just as you and your cousin are linked by being descendants of your grandparents). The fossil record preserves a number of extinct species that inform us about the characteristics of the last common ancestor of reptiles and synapsids, as well as fossil species that are near the base of the reptile and synapsid lines of descent [see e.g., Ruta and Coates (2007) for a sense of the diversity of the species just before this split, and Benton (2005), Kemp (2005), and Prothero (2007) for information on the earliest synapsids and reptiles]. These fossils provide important insights about what characters early members of both groups inherited from their common ancestor, which characters are new features that are unique to one group or the other, and the evolutionary and ecological context in which new characters and species evolved. Scientists use these indirect methods to understand the ancestors of groups of organisms because it is all but impossible to say with certainty whether a particular fossil is definitely an ancestor. However, by studying where various organisms fall on phylogenies and what characteristics the organisms possess, we can get a good sense of what an ancestor was like even if we cannot identify it exactly.
How to Recognize a Synapsid
A final important issue to address in this section is the question of how scientists recognize which animals are part of the group Synapsida. In other words, what does it take to be a synapsid? Once again, tree thinking is critical to answering this question, as are the concepts of definition and diagnosis.
The definition of a named group of organisms describes the limits of membership of that group. With the rise of tree thinking, the definitions of groups have come to focus on patterns of descent from common ancestors, as we saw with the definition of Kielan-Jaworowska et al. (2003) of mammals quoted above. A similar definition for Synapsida could take the form of “all animals more closely related to Homo sapiens than Terrapene carolina (the eastern box turtle).” However, although this definition tells us who is a synapsid (any animal that is more closely related to H. sapiens than to the reptile T. carolina), it does not tell us how to recognize synapsids if we find them as fossils or in the living biota. This is where the concept of diagnosis comes into play: A diagnosis is a list of characters we can see on a specimen that provide insight into the nature of its ancestry (additional information on the distinction of definition and diagnosis in the context of mammal evolution can be found in Rowe 1988; de Queiroz 1994, and Padian and Angielczyk 2007). Typically scientists develop diagnoses for groups by comparing the distribution of characters among different organisms to the relative placement of those organisms on an evolutionary tree. Through this process, they can determine which characters occur in only descendants of a particular common ancestor and thus can be used to identify such descendants. A good example of a diagnostic character that can be used to identify almost any synapsid is the presence of the so-called synapsid temporal opening, an opening on the side of the skull in the vicinity of where the jaw musculature attaches, which can be found (with slight modifications) in synapsids as distinctive as Dimetrodon and living mammals, including humans (Fig. 5). Useful diagnostic characters for living mammals include hair and the presence of mammary glands that allow female mammals to secrete milk to nourish their offspring.
a Photograph of the left side of a fossil Dimetrodon skull (FMNH UC 40) showing the location of the synapsid temporal opening behind the eye socket. b Line drawing of a human skull showing the location of the synapsid temporal opening behind the eye socket. Jaw muscles attach to the skull in the vicinity of the temporal opening, and it is an important character that allows us to identify living and fossil synapsids. Images are not to scale
Now that we understand who synapsids are and where they fall on the tree of life relative to other groups such as reptiles, let us turn to the topic of synapsid diversity and its implications for the evolution of many of the distinctive characters of mammals.
Tree Thinking, Synapsid Diversity, and the Evolution of Mammalian Characters
Because of their close relationship to living mammals and the fact that they provide a record of the evolutionary history of many mammalian characters, much research on non-mammalian synapsids is couched in the framework of mammal origins (e.g., Sidor and Hopson 1998; Sidor 2001, 2003). Similarly, most popular accounts of synapsids make a point of describing the order in which different mammalian characters evolved and the evolutionary steps in the process. Such works are important because they present the parts of synapsid history that are most directly our own and shed light on what our very ancient ancestors were like. However, in the interest of brevity and simplicity, the full diversity of non-mammalian synapsids is rarely addressed. This is unfortunate because although most people realize that living mammals vary from aardvarks and bats to whales and zebras, they are usually unaware of the fascinating array of shapes, sizes, and ways of life that non-mammalian synapsids evolved millions of years before the first mammals appeared. On the occasions when they do encounter some of this diversity, such as during museum visits, their unfamiliarity can lead to confusion about how it pertains to mammal evolution. A complete review of synapsid diversity is beyond the scope of this paper, but a few examples will help to provide a sense of its breadth.
A Selection of Synapsids
Eothyris (Fig. 6a) is one of the most primitive synapsids known (i.e., it falls very close to the base of the synapsid branch of the tetrapod evolutionary tree) and is probably very similar to the common ancestor of all synapsids in many respects. Its teeth suggest that it was a carnivore, but because Eothyris is known only from a skull, it is difficult to say much else about its way of life. The only known specimen of Eothyris was collected in north-central Texas in rocks that are about 275 million years old.
a The right side of the skull and jaw of Eothyris (MCZ 1161) and eothyridid synapsid. Eothyris is probably a good model for the common ancestor of all synapsids. b Skeleton of Edaphosaurus, an edaphosaurid synapsid. Edaphosaurus is one of the oldest known terrestrial vertebrate herbivores. This skeleton is on display at The Field Museum. Images are not to scale
Dimetrodon and Edaphosaurus (Figs. 1, 6b) are two of the best-known and most distinctive early synapsids. Both animals are characterized by large sails on their backs, which are formed by elongated portions of their vertebrae, and which may have evolved as a means for them to more precisely regulate their body temperatures (Bennett 1996). Fascinatingly, evolutionary trees of synapsids suggest that Edaphosaurus and Dimetrodon evolved their sails independently, an example of the process of convergent evolution, where similar selective forces acting on distantly related organisms can result in the evolution of similar structures. Edaphosaurus also is noteworthy because it is one of the oldest known terrestrial vertebrates that is specialized for feeding on plants (Sues and Reisz 1998; Reisz 2006). Dimetrodon and Edaphosaurus are best known from fossil localities in the American southwest, particularly Texas, that range in age between about 300 million and 270 million years in age (Reisz 1986).
Dinocephalians like Anteosaurus and Ulemosaurus (Fig. 7) are more advanced synapsids that are part of the synapsid subgroup called Therapsida (Fig. 4b). The Dinocephalia include both carnivores (like Anteosaurus) and herbivores (like Ulemosaurus), and some members of the group are among the largest non-mammalian synapsids known, reaching sizes similar to living hippos. Although the group’s history is comparatively brief (dinocephalian fossils are known from rocks spanning a narrow time interval between about 269 million to 263 million years ago), it is represented by a diversity of species known from China, Russia, South Africa, Zimbabwe, and Brazil (Rubidge 2005). Some dinocephalians also evolved a number of skull specializations, such as increased bone thickness, that appear to be related to head-butting behaviors (Barghusen 1975).
Dinocephalian specimens. a The left side of the skull and jaw of Anteosaurus (SAM-PK-K11296), a carnivorous dinocephalian. b The left side of the skull of Ulemosaurus (PIN 2207/111), an herbivorous dinocephalian. Note the thickened bone on the upper surface of each skull, which may have been used in head-butting. Images are not to scale
Anomodonts (Fig. 8) are one of the most successful groups of non-mammalian synapsids, and their remains have been discovered on every continent (King 1988, 1992). They are best known from the Middle Permian to the Late Triassic periods of Earth history (about 269 million to about 215 million years ago), although a controversial, fragmentary specimen from Australia hints that they might have survived until the Early Cretaceous, about 110 million years ago (Thulborn and Turner 2005). Anomodonts are noteworthy because they were among the most diverse and abundant herbivores of the Permian and Triassic, making them critical components of terrestrial ecosystems at these times. Most members of the group are toothless and possessed a turtle-like beak in life that they used for chewing their food (Sues and Reisz 1998; Reisz and Sues 2000; Angielczyk 2004; Reisz 2006), although some species retained a pair of large tusks in their upper jaws (Fig. 8a). Anomodonts evolved a wide range of body sizes, with the smallest species being about the size of a living marmot and the largest reaching sizes comparable to hippos or rhinos today. They also evolved diverse lifestyles. For example, the anomodont Cistecephalus (Fig. 8b) and its close relatives appear to have been specialized burrowers (Cox 1972; Cluver 1978), and the anomodont Lystrosaurus has been suggested to have been semi-aquatic, much like living hippos (e.g., Germain and Laurin 2005; Ray et al. 2005).
Anomodont specimens. a Left side of the skull of Delectosaurus (PIN 4644/1). Note the presence of a tusk and the absence of all other teeth. b Skeleton of Cistecephalus (BP/1/4086). Based on features of the front limb and skull, Cistecephalus is thought to have been a specialized burrowing animal (Cluver 1978). c Left side of the skull of Suminia (PIN 2212/62). Suminia differs from most anomodonts by possessing numerous large teeth. However, many features of the skull indicate that it is more closely related to other anomodonts than to any other synapsids. Images not to scale
Gorgonopsians (Fig. 9) are an abundant and widespread group of carnivorous synapsids that are known from fossil localities in Russia and Africa dating to the Middle and Late Permian periods of Earth history (about 269 million to 251 million years ago). Most members of the group were dog- or wolf-like predators, although some species reached sizes comparable to extant polar bears and grizzly bears [Sigogneau-Russell (1989) provides an overview of the diversity of gorgonopsian species; Kemp (1982) and Kemp (2005) give more general introductions to the group]. Gorgonopsians evolved a sabre-tooth appearance with huge canine teeth, similar to what is seen in the more familiar (and much younger) sabre-toothed cat Smilodon. They may have even possessed a specialized jaw joint that allowed them to open their mouths extremely widely, thus enabling them to better employ their canines when attacking prey, but also to shift their jaw forward so that their incisors (i.e., front teeth) could intermesh for more efficient cutting of meat once the prey was subdued (Kemp 1969; Tatarinov 2000; although see Laurin 1998).
Therocephalians (Fig. 10) are another group of mostly carnivorous synapsids that first appeared in the Middle Permian Period of Earth history (about 269 million years ago), although they survived longer than gorgonopsians, finally going extinct in the Middle Triassic (about 243 million years ago). There is debate surrounding the exact relationship of therocephalians to the cynodonts, the non-mammalian synapsids most closely related to mammals. Many workers have treated therocephalians as a distinct group that is descended from a single common ancestor and whose members are more closely related to each other than to any other synapsids (e.g., Hopson and Barghusen 1986; van den Heever 1994; Sidor and Hopson 1998; Rubidge and Sidor 2001; Sidor 2001, 2003). However, some authors have suggested that certain therocephalian taxa may be more closely related to cynodonts than to other therocephalians (Kemp 1972; Abdala 2007; Botha et al. 2007), a hypothesis that would imply that Therocephalia is not a natural group (i.e., it does not consist only of descendants of a single common ancestor; see above). Most therocephalians were carnivores, although smaller species likely were insectivores, and some later-occurring species may have been herbivores or omnivores. They also evolved a variety of body types, including wolf- and weasel-like forms (Fig. 10a, b). The therocephalian Euchambersia (Fig. 10c) is particularly interesting because several features of its skull and teeth are suggestive of it being venomous (Romer 1956; Kemp 1982, 2005; Hotton 1991; Folinsbee et al. 2007).
Therocephalian specimens. a Skeleton of Glanosuchus (SAM-PK-K7809) showing the wolf-like proportions typical of some members of the group. b Left side of the skull and jaw of Mirotenthes (UCMP V3695/40467), a typical small therocephalian. c Left side of the skull of Euchambersia, a therocephalian thought to have been venomous (e.g., Kemp 2005). Note the very different skull proportions compared to Mirotenthes and the presence of pit on the side of the snout that may have housed a venom-secreting gland. Images not to scale
Cynodonts are the non-mammalian synapsids most closely related to mammals. Indeed, mammals are part of the group Cynodontia (i.e., mammals are descendants of the common ancestor of all cynodonts), with cynodont lineages that diverged before the most recent common ancestor of mammals being referred to as non-mammalian cynodonts. The oldest known cynodont was recently discovered in South Africa and was collected in rocks from the early Late Permian Period of Earth history (about 257 million years ago; Botha et al. 2007). However, cynodonts remained relatively rare components of terrestrial faunas until the Triassic period, when they became much more diverse and abundant. Early non-mammalian cynodonts, such as Procynosuchus (Fig. 11a) or Progalesaurus (Fig. 11b), tend to be small insectivorous or carnivorous animals, although in the Triassic, they evolved a wider range of body sizes and lifestyles. Because of their close relationship to mammals, cynodonts continue to be the subject of much research, and this work has done much to elucidate the relationships of the earliest mammals, as well as the origins of many unique mammalian characters (e.g., see reviews in Hopson 1991, 1994; 2001; Kielan-Jaworowska et al. 2003; Kemp 2005; Benton 2005; Prothero 2007).
Non-mammalian cynodont specimens. a Partial skeleton of Procynosuchus (RC 92). b Left side of the skull and jaw of Progalesaurus (SAM-PK-K9954). c Left side of the skull of Diademodon (BSP 1934-VIII-15). Procynosuchus and Progalesaurus are early non-mammalian cynodonts and were carnivores. Diademodon likely was an omnivore or herbivore. Images not to scale
Now that we have seen some of the diversity of non-mammalian synapsids, we can examine how paleontologists use evolutionary trees to organize this diversity and to investigate where in synapsid history different mammalian characters evolved.
Using Tree Thinking to Organize Synapsid Diversity and Track Mammalian Characters
Because evolutionary trees depict patterns of descent from common ancestors, they are an extremely useful way to relate the diverse shapes, sizes, and ways of life observed in a group of animals such as the synapsids to their evolutionary history. In turn, this allows us to accurately determine where in synapsid history particular features of interest evolved, as well as whether they were present in the ancestors of mammals or only in lines of descent that do not lead to mammals.
Recall that when scientists construct a phylogeny, they consider a large number of characters and search for the tree that best explains the evolution of as many of those characters as possible. The resulting trees group species together based on inferred patterns of descent from common ancestors. Usually, the species in particular groups resemble one another for the simple reason that they inherited many of their similarities from common ancestors. However, evolutionary trees often contain some surprises as well. For example, detailed phylogenetic research indicates that Edaphosaurus, which falls in the group called “edaphosaurids” in Fig. 4b, and Dimetrodon, which falls in the group called “sphenacodontids” in Fig. 4b, do not share a common ancestor that possessed a sail on its back, despite the fact that both Edaphosaurus and Dimetrodon have sails on their backs. By grouping organisms together based on patterns of descent inferred from a large number of characters and not just a few potentially superficial similarities or differences, phylogenies provide a framework for asking and answering deeper questions about diversity. For example, do all descendants of a common ancestor share a similar way of life, such as being terrestrial carnivores, or do some evolve to adapt in other ecologies, such as an aquatic herbivore? Is there a correlation between the number of species in a group and the diversity of shapes, sizes, and ecologies found in the group? Do some lines of descent from a common ancestor display higher rates of speciation than others? Do superficially similar animals, such as Dimetrodon and Edaphosaurus, share a recent common ancestor or did they evolve their similarities independently?
In addition to their utility in studies of diversity, phylogenies also can provide great insight into the evolutionary history of individual characters. By “mapping” a character of interest onto a phylogeny, we can not only see which species possess the character but also make inferences about where in the history of the group the character evolved and whether it is likely that the character evolved only once or multiple times [reviews of methods used for mapping the evolution of characters on evolutionary trees can be found in Brooks and McLennan (1991) and Garland et al. (2005)]. The evolution of a bony secondary palate in different groups of synapsids provides a useful example of how tree thinking can elucidate the history of a character.
All living mammals possess a bony secondary palate, a sheet of bone that separates the mouth from the nasal cavity. A number of hypotheses have been put forward for the function of the secondary palate, including better respiratory function while chewing, perhaps in the context of increased metabolic rates (McNab 1978; Hillenius 2004); generation of negative pressure in the mouth cavity while suckling (Maier et al. 1996; Maier 1999); or increasing the mechanical strength of the snout (Thomasson and Russell 1986). It is also possible that more than one of these factors represents the selective pressure involved in the palate’s evolution. The oldest fossil mammals also possess secondary palates (Kielan-Jaworowska et al. 2003), but if we look among non-mammalian synapsids, we find a mixture of animals that lack or possess secondary palates (Fig. 12a). Determining when in synapsid history this feature first evolved and whether it might have evolved more than once are potentially daunting tasks, but the situation becomes much clearer when we map the character onto an evolutionary tree.
Bony secondary palate evolution in synapsids. a Drawings showing the bottom surface of the snout in four synapsids, one lacking a bony secondary palate, the others possessing a bony secondary palate. Note that the internal nostril (the opening through which air passes as it moves from the nasal cavity to the trachea and lungs, highlighted with gray shading) is much further back on the skull in the synapsids with a secondary palate, effectively isolating the path air takes as it enters the body from the mouth. In contrast, the forward position of the internal nostrils in the synapsid lacking a secondary palate means that air must pass through the mouth on its way to the lungs. b Simplified phylogeny of synapsids showing the distribution of bony secondary palates among major groups, as well as comparative drawings of the snouts of various synapsids (internal nostrils highlighted with gray shading). Note that the distribution of secondary palates on this phylogeny implies that the character evolved three separate times in synapsid history. Also note that some synapsids with incomplete secondary palates, such as Ictidosuchoides and Procynosuchus, help to document the evolution of this character (see text for details). Groups shown in bold include living representatives. Drawings were redrawn and modified from Efremov (1940), Romer and Price (1940), Kemp (1969), Hopson and Barghusen (1986), Rybczynski (2000), Angielczyk and Kurkin (2003), Maddin et al. (2008)
The first step in mapping the character onto the tree is to note which of the animals listed at the tips of the branches possessed a bony secondary palate. The results of this step are shown in Fig. 12b. Now, we will need to reconstruct whether the common ancestors of different groups also possessed a secondary palate. There are a number of ways to do this, but for simplicity, we will use a method called parsimony, which seeks to minimize the number of evolutionary changes that are necessary to explain the distribution of the character on the tree. The results of this step reveal an interesting pattern: A bony secondary palate evolved multiple times in synapsids (Fig. 12b). The character first evolved in the common ancestor of advanced anomodonts like Delectosaurus and Cistecephalus. It evolved a second time in the common ancestor of advanced therocephalians such as Bauria and Ericiolacerta. Finally, a bony secondary palate evolved a third time early in cynodont history. We prefer this scenario to others because it requires the fewest evolutionary changes. For instance, if we hypothesized that a secondary palate evolved in the common ancestor of therocephalians and cynodonts, our tree implies that this feature would need to have been lost in several therocephalian subgroups, necessitating four evolutionary changes, as opposed to the three required by our preferred hypothesis. Moreover, the pattern indicates that mammals inherited their bony secondary palate from a cynodont ancestor, whereas the secondary palates of anomodonts and therocephalians represent separate evolutionary innovations that are restricted to those groups, and thus are not directly relevant to the specific history of mammals. Finally, if we consider animals such as Procynosuchus or Ictidosuchoides, we find that although they lack a complete secondary palate, their internal nostrils are partially covered over by an incomplete secondary palate. These forms and their position on the phylogeny are important because they help to document the evolutionary transition between synapsids lacking secondary palates and those with complete secondary palates.
This tree thinking approach can be applied to any character and is very useful for answering a variety of evolutionary questions. For instance, to return to the question of whether our ancestors had a sail like Dimetrodon, character mapping implies that the answer is no. Hypothesizing that the common ancestor of mammals and Dimetrodon possessed a sail requires two evolutionary changes: one change to the sailed condition and a second change to the non-sailed condition in a more recent ancestor of mammals. In contrast, hypothesizing that a sail evolved in the immediate ancestor of Dimetrodon and its closest relatives and not in the common ancestor of Dimetrodon and mammals requires only one evolutionary change (i.e., the evolution of a sail in Dimetrodon’s ancestor; no change to the non-sailed condition is needed). Character mapping also can inform us about the order in which different characters evolved. For example, the method indicates that the novel jaw muscle arrangement found in mammals (including the presence of a masseter muscle) evolved before two of the tiny middle ear bones, which were formerly part of the lower jaw, were fully incorporated into the hearing system (e.g., Kemp 2005).
Conclusions
The evolution of mammals from non-mammalian synapsid ancestors is a significant topic for anyone interested in biology or paleontology. Not only is the process recorded in great detail in the fossil record, giving us much insight into the order in which characters changed and the functional and ecological contexts in which those changes occurred, but it is also part of our history. Because non-mammalian synapsids are our relatives, learning about them helps to answer the question of “where did we come from?” as much as studying fossil hominids or family genealogies. However, confusion often persists because of outdated ways of thinking about the topic.
Tree thinking is an excellent solution to the problem because evolutionary trees provide an excellent framework for asking and answering evolutionary questions and are now a common tool used by biologists and paleontologists. For example, because evolutionary trees depict patterns of descent from common ancestors, they are very useful for understanding the relationship of non-mammalian synapsids to mammals (they are all descendants of a common ancestor), as well as how synapsids are related to reptiles (they are descendants of a common ancestor, but reptiles and synapsids are separate lines of descent). Similarly, phylogenies are very useful for organizing patterns of diversity in large groups, such as the Synapsida, and when combined with techniques such as character mapping, they can provide much insight into the evolution of different characters.
So, when you are next confronted by non-mammalian synapsids, whether on television, at a museum, or in a book on prehistoric life, take a moment to use your tree thinking skills. They will help you to remember why Dimetrodon is not a dinosaur, why our ancestors did not have a sail like that of Edaphosaurus, and how scientists reconstruct the evolutionary history of mammalian characters.
References
Abdala F. Redescription of Platycraniellus elegans (therapsida, Cynodontia) from the Lower Triassic of South Africa and the cladistic relationships of Eutheriodontia. Palaeontology 2007;50:591–618. doi:10.1111/j.1475-4983.2007.00646.x.
Anderson JS, Reisz RR, Scott D, Fröbisch ND, Sumida SS. A stem batrachian from the Early Permian of Texas and the origin of frogs and salamanders. Nature 2008;453:515–8. doi:10.1038/nature06865.
Angielczyk KD. Phylogenetic evidence for and implications of a dual origin of propaliny in anomodont therapsids (Synapsida). Paleobiology 2004;30:268–296.
Angielczyk KD, Kurkin AA. Phylogenetic analysis of Russian Permian dicynodonts (Therapsida: Anomodontia): implications for Permian biostratigraphy and Pangaean biogeography. Zool J Linn Soc 2003;139:157–212. doi:10.1046/j.1096-3642.2003.00081.x.
Barghusen HR. A review of fighting adaptations in dinocephalians (Reptilia, Therapsida). Paleobiology 1975;1:295–311.
Bennett SC. Aerodynamics and thermoregulatory function of the dorsal sail of Edaphosaurus. Paleobiology 1996;22:496–505.
Benton MJ. Vertebrate palaeontology. 3rd edn. Oxford, UK: Blackwell; 2005.
Botha J, Abdala F, Smith R. The oldest cynodont: new clues on the origin and early diversification of the Cynodontia. Zool J Linn Soc 2007;149:477–92.
Brooks DR, McLennan DA. Phylogeny, ecology, and behavior: a research program in comparative biology. Chicago: University of Chicago Press; 1991.
Cantino PD, de Queiroz K. International code of phylogenetic nomenclature. http://www.ohiou.edu/phylocode (2007).
Carroll RL. Problems of the origin of reptiles. Biol Rev Camb Philos Soc 1969a;44:393–432. doi:10.1111/j.1469-185X.1969.tb01218.x.
Carroll RL. Origin of reptiles. In: Gans C, editor. Biology of the Reptilia. Volume 1. London: Academic; 1969b. p. 1–44.
Carroll RL. The ancestry of reptiles. Philos Trans R Soc Lond Ser B 1970;257:367–08.
Chiappe LM. Glorified dinosaurs: the origins and early evolution of dinosaurs. Hoboken: Wiley; 2007.
Cluver MA. The skeleton of the mammal-like reptile Cistecephalus with evidence for a fossorial mode of life. Ann S Afr Mus 1978;76:213–46.
Cox CB. A new digging dicynodont from the Upper Permian of Tanzania. In: Joysey KA, Kemp TS, editors. Studies in vertebrate evolution. Edinburgh, UK: Oliver and Boyd; 1972. p. 173–89.
Cope ED. The theromorphous Reptilia. Am Nat 1878;12:829–30.
Darwin C. On the origin of species by means of natural selection, or the preservation of favoured races in the struggle for life. London: John Murray; 1859.
de Queiroz K. Replacement of an essentialistic perspective on taxonomic definitions as exemplified by the definition of “Mammalia.”. Syst Biol 1994;43:497–510. doi:10.2307/2413548.
Efremov IA. Ulemosaurus svijagensis Riab., ein Dinocephale aus demn Ablagerungen des Perm der USSR. Nova Acta Leopold 1940;9:155–205.
Felsenstein J. Inferring phylogenies. Sunderland: Sinauer; 2004.
Folinsbee K, Müller J, Reisz RR. Canine grooves: morphology, function, and relevance to venom. J Vertebr Paleontology 2007;27:547–51. doi:10.1671/0272-4634(2007)27[547:CGMFAR]2.0.CO;2.
Garland T, Bennett AF, Rezende EL. Phylogenetic approaches in comparative physiology. J Exp Biol 2005;208:3015–35. doi:10.1242/jeb.01745.
Germain D, Laurin M. Microanatomy of the radius and lifestyle in amniotes (Vertebrata, Tetrapoda). Zool Scr 2005;34:335–50. doi:10.1111/j.1463-6409.2005.00198.x.
Gregory TR. Understanding evolutionary trees. Evolution: Education and Outreach 2008;121–37.
Hillenius WJ. Turbinates in therapsids: evidence for Late Permian origins of mammalian endothermy. Evolution Int J Org Evolution 2004;48:207–29. doi:10.2307/2410089.
Hopson JA. The mammal-like reptiles: a study of transitional fossils. Am Biol Teach 1987;49:16–26.
Hopson JA. Systematics of the nonmammalian Synapsida and implications for patterns of evolution in snyapsids. In: Schultze HP, Trueb L, editors. Origins of the higher groups of tetrapods. Ithaca, NY: Comstock; 1991. p. 635–93.
Hopson JA. Synapsid evolution and the radiation of non-eutherian mammals. In: Prothero DR, Schoch RM, editors. Major features of vertebrate evolution. Short Courses in Paleontology 1994;7:190–219.
Hopson JA. Origin of mammals. In: Briggs DEG, Crowther PR, editors. Palaeobiology II. Oxford, UK: Blackwell Science; 2001. p. 88–94.
Hopson JA, Barghusen HR. An analysis of therapsid relationships. In: Hotton N, MacLean PD, Roth JJ, Roth EC, editors. The ecology and biology of mammal-like reptiles. Washington D.C.: Smithsonian Institution Press; 1986. p. 83–106.
Hotton N. The nature and diversity of synapsids: prologue to the origin of mammals. In: Schultze HP, Trueb L, editors. Origins of the higher groups of tetrapods. Ithaca, NY: Comstock; 1991. p. 598–634.
Huelsenbeck JP, Ronquist F. Bayesian analysis of molecular evolution using MrBayes. In: Nielsen R, editor. Statistical methods in molecular evolution. New York: Springer; 2005. p. 183–232.
Hull DL. Science as a process. Chicago: University of Chicago Press; 1988.
Kemp TS. On the functional morphology of the gorgonopsid skull. Philos Trans R Soc Lond B Biol Sci 1969;256:1–83. doi:10.1098/rstb.1969.0036.
Kemp TS. Whaitsiid Therocephalia and the origin of cynodonts. Philos Trans R Soc Lond B Biol Sci 1972;264:1–54. doi:10.1098/rstb.1972.0008.
Kemp TS. Mammal-like reptiles and the origin of mammals. New York: Academic; 1982.
Kemp TS. The origin and evolution of mammals. Oxford, UK: Oxford University Press; 2005.
Kielan-Jaworowska Z, Cifelli RL, Luo ZX. Mammals from the age of dinosaurs. New York: Columbia University Press; 2003.
King GM. Anomodontia. Handbuch der Paläoherpetologie 17C. Stuttgart: Gustav Fischer Verlag; 1988.
King GM. The palaeobiogeography of Permian anomodonts. Terra Nova 1992;4:633–40. doi:10.1111/j.1365-3121.1992.tb00612.x.
Kitching IJ, Forey PL, Humphries CJ, Williams DM. Cladistics: the theory and practice of parsimony analysis. Syst Assoc Publ 1998;11:1–228.
Laurin M. New data on the cranial anatomy of Lycaenops (Synapsida, Gorgonopsidae), and reflections on the possible presence of streptostyly in gorgonopsians. J Vertebr Paleontology 1998;18:765–76.
MacFadden BJ. Fossil horses: systematics, paleobiology, and evolution of the family Equidae. Cambridge, UK: Cambridge University Press; 1992.
Maddin HC, Sidor CA, Reisz RR. Cranial anatomy of Ennatosaurus tecton (Synapsida: Caseidae) from the Middle Permian of Russia and the evolutionary relationships of Caseidae. J Vertebr Paleontology 2008;28:160–80. doi:10.1671/0272-4634(2008)28[160:CAOETS]2.0.CO;2.
Maier W. On the evolutionary biology of early mammals—with methodological remarks on the interaction between ontogenetic adaptation and phylogenetic transformation. Zool Anz 1999;238:55–74.
Maier W, van den Heever J, Durand J. New therapsid specimens and the origin of the secondary hard and soft palate of mammals. J Zoological Syst Evol Res 1996;34:9–19.
Martin RA. Missing links. Sudbury: Jones and Bartlett; 2004.
McNab BK. The evolution of endothermy in the phylogeny of mammals. Am Nat 1978;112:1–21. doi:10.1086/283249.
O’Hara RJ. Homage to Clio, or, toward an historical philosophy of evolutionary biology. Syst Zool 1988;37:142–55. doi:10.2307/2992272.
O’Hara RJ. Population thinking and tree thinking in systematics. Zool Scr 1997;26:323–9. doi:10.1111/j.1463-6409.1997.tb00422.x.
Padian K, Angielczyk KD. “Transitional forms” versus transitional features. In: Petto AJ, Godfrey LR, editors. Scientists confront intelligent design and creationism. New York: Norton; 2007. p. 197–230.
Prothero DR. Evolution: what the fossils say and why it matters. New York: Columbia University Press; 2007.
Ray S, Chinsamy A, Bandyopadhyay S. Lystrosaurus murrayi (Therapsida, Dicynodontia): bone histology, growth, and lifestyle adaptations. Palaeontology 2005;48:1169–85. doi:10.1111/j.1475-4983.2005.00513.x.
Reisz RR. Pelycosauria. Handbuch der Paläoherpetologie 17A. Stuttgart: Gustav Fischer Verlag; 1986.
Reisz RR. Origin of dental occlusion in tetrapods: signal for terrestrial vertebrate evolution? J Exp Zool 2006;306B:261–77, Molecular Development and Evolution. doi:10.1002/jez.b.21115.
Reisz RR, Sues HD. Herbivory in late Paleozoic and Triassic terrestrial vertebrates. In: Sues HD, editor. Evolution of herbivory in terrestrial vertebrates. Cambridge, UK: Cambridge University Press; 2000. p. 9–41.
Romer AS. Osteology of the reptiles. Chicago: University of Chicago Pressp; 1956.
Romer AS, Price LI. Review of the Pelycosauria. Geological Society of America Special Paper 1940;28:1–538.
Rowe T. Definition, diagnosis, and the origin of Mammalia. J Vertebr Paleontology 1988;8:241–64.
Rubidge BS. Re-uniting lost continents—fossil reptiles from the ancient Karoo and their wanderlust. S Afr J Geol 2005;108:135–72. doi:10.2113/108.1.135.
Rubidge BS, Sidor CA. Evolutionary patterns among Permo-Triassic synapsids. Annu Rev Ecol Syst 2001;32:449–80. doi:10.1146/annurev.ecolsys.32.081501.114113.
Ruta M, Coates MI. Dates, nodes, and character conflict: addressing the lissamphibian origin problem. J Syst Palaeontology 2007;5:69–122. doi:10.1017/S1477201906002008.
Rybczynski N. Cranial anatomy and phylogenetic position of Suminia getmanovi, a basal anomodont (Amniota: Therapsida) from the Late Permian of eastern Europe. Zool J Linn Soc 2000;130:329–73. doi:10.1111/j.1096-3642.2000.tb01634.x.
Sidor CA. Simplification as a trend in synapsid cranial evolution. Evolution Int J Org Evolution 2001;55:1419–42.
Sidor CA. Evolutionary trends and the origin of the mammalian lower jaw. Paleobiology 2003;20:605–40. doi:10.1666/0094-8373(2003)029%3C0605:ETATOO%3E2.0.CO;2.
Sidor CA, Hopson JA. Ghost lineages and “mammalness”: assessing the temporal pattern of character acquisition in the Synapsida. Paleobiology 1998;24:254–73.
Sigogneau-Russell D. Theriodontia I. Handbuch der Paläoherpetologie 17B I. Stuttgart: Gustav Fischer Verlag; 1989.
Simpson GG. Diagnosis of the classes Reptilia and Mammalia. Evolution Int J Org Evolution 1960;14:388–92. doi:10.2307/2405982.
Sues HD, Reisz RR. Origins and early evolution of herbivory in tetrapods. Trends Ecol Evol 1998;13:141–5. doi:10.1016/S0169-5347(97)01257-3.
Tatarinov LP. Problems of the morphology of the theriodonts (Reptilia). Russ J Herpetol 2000;7:29–40.
Thomason JJ, Russell AP. Mechanical factors in the evolution of the mammalian secondary palate: a theoretical analysis. J Morphol 1986;189:199–213. doi:10.1002/jmor.1051890210.
Thulborn T, Turner S. The last dicynodont: an Australian Cretaceous relict. Proc R Soc Lond B Biol Sci 2005;270:985–93. doi:10.1098/rspb.2002.2296.
van den Heever J. The cranial anatomy of the early Therocephalia (Amniota: Therapsida). Ann Univ Stellenbosch 1994;1994:1–59.
Acknowledgments
Thanks are due to Don Prothero for organizing this special issue of Evolution: Education and Outreach and for inviting me to contribute to it. I also thank curators, collections managers, and friends at the Bayerische Staatssammlung für Paläontologie und historische Geologie (BSP), the Bernard Price Institute for Palaeontological Research (BP), The Field Museum (FMNH), the Iziko South African Museum (SAM), the Museum of Comparative Zoology (MCZ), the Palaeontological Institute (PIN), the Rubidge Collection (RC), and the University of California Museum of Paleontology (UCMP) for their assistance with the visits on which I took the photos in Figs. 5, 6, 7, 8, 9, 10, and 11. Christian Kammerer graciously allowed me to use the photos in Figs. 7b, 10c, and 11b. Audrey Aronowsky and Scott Buhrman commented on drafts of the manuscript. Christian Sidor and two anonymous reviewers also provided helpful comments and suggestions.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License ( https://creativecommons.org/licenses/by-nc/2.0 ), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Angielczyk, K.D. Dimetrodon Is Not a Dinosaur: Using Tree Thinking to Understand the Ancient Relatives of Mammals and their Evolution. Evo Edu Outreach 2, 257–271 (2009). https://doi.org/10.1007/s12052-009-0117-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12052-009-0117-4