Abstract
Gerstmann-Sträussler-Scheinker (GSS) syndrome is a fatal autosomal dominant neurodegenerative prionopathy clinically characterized by ataxia, spastic paraparesis, extrapyramidal signs and dementia. In some GSS familiar cases carrying point mutations in the PRNP gene, patients also showed comorbid tauopathy leading to mixed pathologies. In this study we developed an induced pluripotent stem (iPS) cell model derived from fibroblasts of a GSS patient harboring the Y218N PRNP mutation, as well as an age-matched healthy control. This particular PRNP mutation is unique with very few described cases. One of the cases presented neurofibrillary degeneration with relevant Tau hyperphosphorylation. Y218N iPS-derived cultures showed relevant astrogliosis, increased phospho-Tau, altered microtubule-associated transport and cell death. However, they failed to generate proteinase K-resistant prion. In this study we set out to test, for the first time, whether iPS cell-derived neurons could be used to investigate the appearance of disease-related phenotypes (i.e, tauopathy) identified in the GSS patient.
Similar content being viewed by others
Introduction
Biomedical research on neurodegenerative diseases with low prevalence in humans relies on the possibility of analyzing brain samples only at very late stages of the disease. Thus, our view of the biochemical or molecular changes during the disease is partial. This drawback steadily increases with a faster neurodegenerative progression speed (e.g., in prionopathies [1] or rapid Alzheimer’s disease [2]). This is also the case for most sporadic taupathies and in most cases of frontotemporal lobar degeneration (FTLD) displaying neurofibrillary degeneration [3, 4]. This limitation impedes the study of early onset changes in asymptomatic patients, making it impossible to investigate illness evolution, therefore hampering biochemical/molecular studies and drug discovery [5].
Gerstmann-Sträussler-Scheinker syndrome (GSS) is a rare autosomal dominant neurodegenerative prionopathy clinically characterized by a wide spectrum of manifestations including but not limited to ataxia, spastic paraparesis, extrapyramidal signs and dementia [6, 5]. Most GSS patients have the P102L mutation in the cellular prion protein (PrPC) gene (PRNP) located in the short arm of chromosome 20 [5]. Cases of rapid progressive forms of GSS are rare [7] with an average duration after clinical diagnosis of 5–6 years (from 6 months to 13 years) [8, 5]. Histopathological examination of post-mortem GSS brains has revealed abnormal misfolded prion (PrP) aggregates in the form of unicentric and multicentric deposits in the cerebellum and cortical gray matter [5]. In addition, western blot analysis of aggregated PrP is distinguished by the presence of truncated protein fragments ranging between 6 and 10 KDa and a variable number of bands of higher molecular weight [9]. Parallel to this particular PrP deposition, pathological features characteristic of other neurodegenerative diseases such us parkinsonism or Alzheimer’s disease have been observed in some GSS patients [5]. Indeed, an increase in hyperphosphorylated Tau is frequently observed in the pathological analysis of brains from GSS patients carrying PRNP mutations P102L [10], P105L [11], A117V [12], V176G [13], F198S [14, 15], Q217R [16, 15] and Y218N [17]. Although it has been shown that PrPC with the P102L mutation display an increased binding to Tau [18], the role of these point mutations in the development of neurofibrillary degeneration is unknown. Nevertheless, in some P102L GSS cases with increased levels of p-Tau, the distribution of p-Tau tangles close to PrP deposits suggesting an active participation of PrP in the generation of p-Tau [10].
Due to the above-mentioned restrictions in this study we explored the usefulness of an induced pluripotent stem (iPS) cell model derived from somatic cells from a GSS patient. iPS cell technology is a tool for basic and translational research through generating in vitro models of disease-relevant cells reprogrammed directly from patients [19–21]. This approach has been shown to be particularly useful in the case of congenital or early-onset monogenic diseases [22] as well as other neurodegenerative diseases [23]. iPS cells have been generated from patients with Alzheimer’s [24], Parkinson’s [25, 26], Hungtinton’s [27] diseases as well as FTLD [28], Amyotrophic Lateral Sclerosis (ALS) [29] and several others. However, there are no reports of iPS cell lines derived from patients with familial prionopathies.
In this study, we generated iPS cells from dermal fibroblasts of a family member of the Y218N GSS patient described by Alzualde and colleagues [17] and differentiated them into neurons using two previously published procedures [30, 31]. To date, very few individuals have been reported carrying this mutation [17, 32]. We were interested in this familiar since the Y218N patient displayed widespread neurofibrillary degeneration in the brain [17]. Results determined that although differentiated Y218N iPS cells were not able to spontaneously generate or propagate human prions, Y218N-derived cultures showed relevant astrogliosis and cell death. In addition, differentiated Y218N-derived neurons displayed high levels of p-Tau, thus recapitulating most of the neuropathological features reported in the patient [17].
Material and Methods
Case Patient
The index case and the younger sister was examined at the Cognitive Disorders Unit at Donostia Hospital. The clinical report of the family and the Y218N patient can be seen in [17]. Dermal fibroblasts were obtained from the younger sister of the Y218N patient (54 years old in 2010) after having made complaints of poor concentration, apathy, emotional lability, and increasing difficulties in planning and executing actions. She had previously been diagnosed with and treated for a depressive illness, and the neuropsychological examination revealed slight memory dysfunction in retrieval, language impairment followed by anomia with preserved verbal comprehension, and executive dysfunction. The Mini Mental State Examination (MMSE) score was 23/30. Magnetic resonance imaging showed slight frontotemporal atrophy and EEG analysis revealed intermittent frontotemporal delay. An additional EEG, 6 months later, showed slow background activity in the patient, with intermittent delta waves in the left hemisphere. 10 months after onset, she had language difficulties, with impairment in semantic knowledge, and MMSE score dropped to 13/30.
Generation of iPS Cells
All experiments were performed under the guidelines and protocols of the Ethical Committee for Animal Experimentation (CEEA) of the University of Barcelona. All procedures adhered to internal and EU guidelines for research involving derivation of pluripotent cell lines. All subjects gave informed consent for the study using forms approved by the Ethical Committee on the Use of Human Subjects in Research at Hospital Donostia in San Sebastián, Spain. Generation of iPSC lines was approved by the Advisory Committee for Human Tissue and Cell Donation and Use, by the Commission on Guarantees concerning the Donation and Use of Human Tissues and Cells of the Carlos III Health Institute, Madrid, Spain (Ref: 589, 1/21/2015). All procedures were done in accordance with institutional guidelines and the cell lines have been (or will be) deposited at the Banco Nacional de Lineas Celulares (BNLC, ISCIII) following the Spanish legislation. Fibroblasts from a healthy individual and from the Y218N GSS patient were infected with retroviruses carrying human cDNA coding for KLF4, SOX2, and OCT4, with or without the addition of c-MYC as previously described [33]. Fibroblasts were maintained in DMEM (Sigma) supplemented with 10% FBS (Life Technologies) and 1% Pen/Strep solution (Life Technologies) before infection. After infection, fibroblasts were plated on irradiated human foreskin fibroblasts (HFF, ATCC) and maintained with hESC medium for 4–12 weeks until iPS cell colonies appeared. Several clones from each cell line were obtained and validated. Y218N patient (FH10) and parallel control (FHB1) iPS cell clones were analyzed in details (see below).
Characterization of iPS Cell Lines
AP staining was performed using the Alkaline Phosphatase Blue Membrane Substrate Solution (Sigma). For immunocytochemistry, cells were grown on HFF feeder layers for 6–10 days and then fixed in 4% PFA for 10 min. After embryonic bodies (EB) formation, differentiation into the 3 germ layers was performed. For endoderm, EBs were plated on 6-well plates treated with Matrigel (BD Biosciences) for 1 h at room temperature, and maintained for 28 days with EB medium. The same procedure was used for mesoderm, but instead using EB medium with 0.5 mM of ascorbic acid. For ectoderm differentiation, EBs were maintained in suspension for 10 days with Neurobasal medium containing N2, B27 and FGF2 (N2B27 medium), prepared as previously described [26]. EBs were then plated on 6-well Matrigel-coated plates and maintained for 21 days with N2B27 medium without FGF supplementation. Differentiated cells were fixed in 4% PFA for 10 min. For nuclear DAPI staining (Invitrogen), 0.5 μg/ml was used. The slides were mounted with PVA:DABCO mounting medium. Images were acquired with an SP2 confocal system (Leica) and analyzed with ImageJ™ software. RT-qPCR analysis was performed as previously described [26]. All results were normalized to the average expression of Glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Transcript-specific primers used are shown in Supplementary Table 1.
For karyotyping, iPS cells were grown on Matrigel and treated with colcemid (Life Technologies) at a final concentration of 20 ng/ml. Karyotyping analysis was carried out by Prenatal Genetics S.L. (Barcelona). For promoter methylation, testing reprogramming gene integration and sequencing to confirm that patient iPS cells were carrying mutations in PRNP gene, DNA was isolated using the QIAamp DNA Mini Kit (QIAGEN) following the manufacturer’s instructions. Bisulfit conversion of the promoters was carried out using the Methylamp DNA modification kit (Epigentek). Five clones of each promoter for each cell line were analyzed by sequencing. The primers used for testing gene integration are shown in Supplementary Table 1.
iPS Cell Differentiation to Neural Cells
In this study, two protocols were used to differentiate the iPS cells. In the first protocol, iPS cell colonies were mechanically passaged onto Matrigel-coated 6-well plates. 24 h later the mTeSR™ was replaced by DDM neural induction medium [34, 35] with the addition of the ALK inhibitor, SB431542 at 10 mM for 4 days (Tocris) and the BMP inhibitor, LDN-193189 at 100 nM (Miltenyi Biotech) for 12 days. Cells were propagated in this medium for 3 weeks. At about 24 days in vitro, cells were dissociated and plated onto wells coated with poly-L-lysine (33.3 mg/ml, BD) and laminin (3.3 mg/ml, BD), and the medium was changed to N2B27 medium. For immunofluorescence, neurons were dissociated with Accutase (Sigma) and replated on glass coverslips coated with poly-L-lysine and laminin. Characterization was done as previously described [36].
In the second procedure, spherical neural masses (SNMs) were obtained as previously described [37]. SNMs were fixed in 4% phosphate buffered paraformaldehyde (PFA) for 2 h and characterized by immunostaining. For nuclear DAPI staining (Invitrogen), 5 μg/ml was used. Mounting medium and imaging analysis were performed for in vitro differentiation testing. SNMs obtained from control and Y218N iPS cells, having been maintained in suspension, were then plated on slide-flasks, 6-well plates, 35 mm ∅ plates or 10 mm ∅ plates all previously treated with Matrigel for 1 h at room temperature, and differentiated for 3, 6 or 9 weeks with N2B27 [26], without FGF supplementation to obtain neural cultures. The correct differentiation was assessed by immunostaining. Antibodies used are shown in Supplementary Table 2. For nuclear DAPI staining (Invitrogen), 0.5 μg/ml was used. The slides were mounted with Mowiol mounting medium.
RT-PCR Protocol
Quantitative real time PCR was performed on total RNA extracted with mirVana’s isolation kit (Ambion) from differentiating iPS cells. Purified RNAs were used to generate the corresponding cDNAs, which served as PCR templates for mRNA quantification. Quantitative RT-PCR assays were performed in duplicate on cDNA samples obtained from the retro-transcription reaction diluted 1:20 in 384-well optical plates (Kisker Biotech) using the ABI Prism 7900 HT Sequence Detection System (Applied Biosystems). The reactions were carried out using 20xTaqMan gene expression assays for genes and 2xTaqMan Universal PCR Master Mix (Applied Biosystems). The reactions were conducted using the following parameters: 50 °C for 2 min, 95 °C for 10 min, 40 cycles at 95 °C for 15 s and 60 °C for 1 min. The fold change was determined using the eq. 2-ΔΔCT. Primers used in iPS cell differentiation experiments and Tau R3/R4 analysis can be seen in Supplementary Table 3.
Sample Collection and Proteinase K Treatment
Samples of control and Y218N differentiating cultures were collected at several differentiation times and were homogenized in 10% lysis buffer (100 mM NaCl, 10 mM EDTA, 0.5% Nonidet P-40, 0.5% sodium deoxycholate, and 10 mM Tris, pH 7.5). Debris were removed with low-speed centrifugation at 3000×g for 10 min, and the supernatants were collected. To detect the presence of Proteinase K (PK)-resistant PrP in the supernatant, homogenates were digested with a final concentration of 10–50 μg/ml PK at 37 °C for 60 min prior to western blot analysis using 3F4 antibody against PrPC. To evaluate the PK resistance of protein samples from the original Y218N patient [17], type I sporadic Creutzfeldt-Jakob disease (sCJD) and Type II sCJD brain homogenates were also processed in parallel. PK digestion was terminated by adding Laemmli buffer and heating the samples at 100 °C for 10 min.
Western Immunoblot
Samples from different differentiation stages from iPS cells to neuronal cultures were processed for western blot, including human post-mortem samples and control cultured cells. The collected samples were homogenized in (10% wt/vol) of 50 mM Tris-HCl, pH 7.4/150 mM NaCl/0.5% Triton X-100/0.5% Nonidet P-40 and a mixture of proteinase inhibitors. After this, samples were centrifuged at 15,000 x g for 20 min at 4 °C. The resulting supernatant was normalized for protein content using BCA kit (Pierce). Cell extracts containing Laemmli buffer were boiled at 100 °C for 10 min, followed by 8–10% SDS electrophoresis, then electrotransferred to nitrocellulose membranes for 2 h at 4 °C. Membranes were then blocked with 5% not-fat milk in 0.1 M Tris-buffered saline (pH. 7.4) for 2 h and incubated overnight in 0.5% blocking solution containing primary antibodies. After incubation with peroxidase-tagged secondary antibodies (1:2000 diluted), membranes were revealed with ECL-plus chemiluminescence western blot kit (Amershan-Pharmacia Biotech). In our experiments, each nitrocellulose membrane was used to detect p-Tau (AT-8 and PHF1 antibodies), Actin, Tubulin as protein loading controls. A list of the antibodies used in these experiments can be seen in Supplementary Table 2.
Densitometry and Statistical Processing
For quantification, developed films were scanned at 2400 × 2400 dpi (i800 MICROTEK high quality film scanner), and the densitometric analysis was performed using Quantity One Image Software Analysis (Biorad). Statistical analysis of the obtained data (RT-qPCR and Western blot) was performed using Bonferroni post hoc test (Multiple comparison test) using GraphPad Prism 6 (Mac OsX, Grahpad). Data are presented as mean ± standard error of the mean (S.E.M.). Differences between groups were considered statistically significant between **** P < 0.001, *** P < 0.01 and ** P < 0.05.
Immunohistochemistry
Differentiating iPS cell cultures were fixed in 4% PFA at different days in culture and then permeabilized with 0.1% Triton X-100 (Sigma) in 0.1 M PBS. After fixation, and extensive rinsing with 0.1 M PBS, cultures were blocked with 10% FBS in 0.1 M PBS prior to incubation with primary antibodies (see Supplementary Table 2). After incubation with primary antibodies, cells were incubated with the pertinent Alexa Fluor-tagged secondary antibodies (Alexa-488 goat anti-mouse or Alexa-568 goat anti-rabbit) (Invitrogen-Life Technologies). Finally, cells were stained with 0.1 μM DAPI (Sigma) diluted in 0.1 M PBS, mounted on Mowiol, and viewed using an Olympus BX61 fluorescence microscope, Zeiss LSM or a Leica SP5 confocal microscopy.
Corrected Total Cell Fluorescence (CTCF) Measurement
CTCF levels of p-Tau (red channel) and MAP2 (green channel) were measured in 150 (Y218N) and 165 (control) identified neurons after 21 days of differentiation using ImageJ™ software following published instructions http://sciencetechblog.com/2011/05/24/measuring-cell-fluorescence-using-imagej/. See also [38] for details. CTCF values were determined using the following formula. CTCF = Integrated Density – (Area of selected cell x Mean fluorescence of background readings). Statistical analysis of the obtained data was performed using Mann-Whitney U test using GraphPad Prism 6 (Mac OsX, Grahpad). Differences between groups were considered statistically significant between **** P < 0.001.
Mitochondrial Movement Analysis in iPS Cell-Derived Neurons
SNM-derived neurons were incubated after 21 days of differentiation with MitoTracker (Molecular Probes) and filmed using a Leica TCS SP2 confocal microscope (Leica) equipped with a 63× immersion oil objective. Time-lapse series of image stacks composed of 10 images (512 × 512 pixels) were taken every 3 s over 10 min. Movies were generated at 10 frames per second. Forty-two axons were registered and analyzed in each group recorded. In all cases, a mitochondrion was considered motile when it moved more than 0.5 μm during 1 min of recording. Distances and speeds of retrograde and anterograde transport were measured, and no tracking pluging was used. ImageJ™ software was used to quantify mitochondrial movement. For each mitochondrion movement, the minimum displacement and the average over time were plotted. Statistical analysis of the obtained data was performed using Mann-Whitney U test using GraphPad Prism 6 (Mac OsX, Grahpad). Differences between groups were considered statistically significant between *** P < 0.01 and ** P < 0.05.
Infectivity Assay
Brain homogenates (10% in sterile PBS) were made fresh the day of the infection. One aliquot was kept frozen at −80 °C to repeat the exposure 72 h after the first infection, as described [39]. Representative samples were taken to confirm the presence of PK-resistant PrP in the homogenates, following digestion with 50 μg/ml for scrapie (263 K) and Creutzfeldt-Jakob disease (CJD) brains and 12.5 μg/ml for the Y218N brain [40]. Control and Y218N forebrain neuronal cultures were infected at early (30–40) and middle (60–80) differentiation times. Neurons were replated 5–6 days before the experiment. The culture supernatant was replaced by fresh media containing 10% brain homogenate (day 1) and this was repeated 72 h later (day 3). Two days later, fresh medium (without inocula) was added without removing the supernatant. At day 10 post-inocula (dpi) the entire medium was replaced and the cells were washed several times with sterile PBS before adding fresh Neurobasal containing B27 and N2 supplements. Medium was replaced every other day for the first 2 weeks and then twice a week until cells were collected or fixed for analysis, ~2 months later. All experiments were performed in a Biosafety level 3 security laboratory.
Results
Generation and Characterization of Y218N GSS Patient-Specific iPS Cells
Fibroblasts were reprogrammed at early passages (5–7) through the retroviral delivery of SOX2, KLF4, OCT4, and c-MYC to generate up to 5 independent iPS cell lines for each individual (Fig. 1). We selected clones displaying embryonic stem cell-like morphology and positive AP staining (Fig. 1a). 5 clones representing each individual were chosen to be thoroughly characterized and shown to be fully reprogrammed, as judged by demethylation of OCT4 and NANOG promoters (Fig. 1b), the silencing of the reprogramming transgenes (Fig. 1c), activation of endogenous pluripotency-associated factors (Fig. 1c), expression of pluripotency-associated transcription factors and surface markers (Fig. 1d), pluripotent differentiation ability in vitro and/or in vivo (Fig. 1f), and karyotype stability after more than 15 passages (Fig. 1e). Mutation analysis confirmed that iPS cells and their derivatives bore the mutation Y218N present in the patient fibroblasts (Fig. 1g).
Generation and characterization of iPS cells. (a) Control (cell line FHB1) and GSS-Y218N-iPS cell (cell line FH10) stained for AP activity. (b) Bisulphite genomic sequencing of the OCT4 and NANOG promoters showing demethylation in FHB1 and FH10 (Y218N) cell lines. (c) RT-qPCR analyses of the expression levels of retroviral-derived reprogramming factors (transgenic) and endogenous expression levels (endogenous) of the indicated genes in FHB1 (two clones) and Y218N-iPS cells (cell line FH10, 2 clones). (d) Low fluorescence photomicrographs of representative colonies of FHB1 and FH10 (Y218N) stained positive for the pluripotency-associated markers OCT4, NANOG and SOX2 (green), SSEA3, TRA-1-81 and SSEA4 (red). (e) Normal karyotypes of FHB1 and FH10 (Y218N) at passage 20. (f) Immunofluorescence analyses of FHB1 and FH10 (Y218N) iPS cells differentiated in vitro show the potential to generate cell derivatives of all three primary germ cell layers including ectoderm (stained for TUJ1, green), endoderm (stained for α-fetoprotein, green, and FOXA2, red) and mesoderm (stained for smooth muscle actin, SMA, red). (g) Direct sequence of genomic DNA from Control (cell line FHB1) and GSS patient (FH10 (Y218N)) identifying the PRNP Y218N mutation. Scale bars in a, d and f = 50 μm
Late Neuronal Maturation, Increased Reactive Astrogliosis and Absence of PrP Generation in Y218N-Derived iPS Cell Cultures
Control and Y218N-derived iPS cells were differentiated into neural cells using two well-characterized procedures (see Methods). Neural induction was fast and efficient using both protocols (Fig. 2) and the cells sequentially expressed typical markers of neural progenitors, neuroblasts and mature neurons (Fig. 2b, f). In our first approach (Fig. 2a-d), taking into account morphology and marker expression, we established three differentiation stages: early (≈ 60 DIV), middle (≈ 60–120 DIV) and late (≈ 120–210 DIV) (Fig. 2a). Two weeks after neural induction, cultures were composed mainly of neural progenitors co-expressing SOX2 and NESTIN with a few differentiating neuroblasts (class III β-tubulin (TUJ1)-positive). By 4 weeks many neuroblasts and young neurons expressed PAX6 and the vast majority expressed Doublecortin (DCX) and Ubiquitin-protein ligase E3A (UBE3A) proteins (Fig. 2b). From the third month onwards, differentiating neurons expressed the mature post-mitotic neuronal marker NeuN (RBFOX3). In parallel and as also reported in vivo [41], PrPC expression increased progressively over time during differentiation (Fig. 2c–d).
Neural differentiation of FHB1 and FH10 ( Y218N ) iPS cells. IPS cells from control (FHB1) and Y218N (FH10) GSS patient were differentiated using two procedures (a and e) (see Methods for details). (b) Low power photomicrographs of representative colonies of FHB1 and FH10 (Y218N) stained positive for SOX2, Nestin, DCX, TUJ1, PAX6, UBE3A antigens at different stages of maturation. (c) Western blot characterization of PrPC expression in differentiating iPS cell cultures. (d) Example of the Western blot experiments illustrating the absence of PK-resistant PrPC in FH10 (Y218N) cultures. (e) Low power photomicrographs of representative colonies of FHB1 and FH10 (Y218N) stained positive for Nestin, Ki67, SOX2, TUJ1, PAX6 and GFAP antigens. (g) Western blot characterization of PrPC expression in iPS cells (passage 20) and SNMs (passage 3). (h) Western blots illustrating the absence of PK-resistant prion in FHB1 and FH10 (Y218N) in brain extracts from the GSS patient and two CJD (Type I and II) samples. Scale bars in b and f = 50 μm
In the second protocol (Fig. 2e-h), control and Y218N-iPS cells were also differentiated to pure masses of neural precursors using a previously described protocol that involves the formation of EBs and the culture of neural precursor cells to form SNMs, whom can be expanded and differentiated into mature neurons after several weeks using neuronal induction medium (Fig. 2e). In these conditions, SNMs derived from control and Y218N-iPS cell lines homogeneously expressed neural progenitor markers such as PAX6, NESTIN, and SOX2, as well as the proliferation marker Ki67 (Fig. 2f). Furthermore, when iPS cell-derived SNMs were cultured in neuronal induction medium supplemented with N2 and B27, differentiation into mature neurons was evident within 3 to 5 weeks (Fig. 2f). After about 3 weeks in neuronal medium, the cultures formed dense MAP2 and TUJ1-positive neuronal networks (Fig. 2f) in presence of astroglial cells (not shown). No mixed genotypes (GFAP + MAP2 or TUJ1 double labeled cells) were observed. As observed in the first approach, PrPC was clearly present throughout neural differentiation (Fig. 2c,g, Supplementary Fig. 1). However, no detectable PK-resistant PrP was observed in protein extracts treated with the enzyme in the Y218N and control-derived neurons generated with either protocol (Fig. 2d,h), in contrast to brain extracts from Y218N GSS or Type 1–2 CJD patients (Fig. 2h, Supplementary Fig. 2).
We next examined the transcriptional profile of neural cultures determined by RT-qPCR from early to late culture stages (Fig. 3a) and observed significant differences between control and Y218N cultures, particularly at the late stage (>120 days). While there were no differences in early progenitor markers such as NES and SOX2, which showed a similar time-dependent downregulation in both genotypes, a few neuronal transcripts were lower in the Y218N cultures early on, like MAP2 and CALB. At the late stage (>120 days), there was a robust increase in GFAP mRNA and a concomitant decrease in mature neuronal markers including MAPT and VGLUT1 mRNAs in Y218N cultures compared to controls. Next, these mRNA changes were checked by immunohistochemistry and cell counts (Fig. 3b, d). Cell counts revealed that the relative percentage of DCX and GFAP expressing cells was not significantly different between control and Y218N cultures (data not shown). Thus, the transcriptional increase in GFAP expression most likely due to a greater expression in reactive astroglial cells (Fig. 3c). Indeed, detailed analysis of immunoreacted cells revealed high content of GFAP forming thick fascicles in hypertrophic reactive astroglial cells at the late stage of Y218N cultures (Fig. 3c). Lastly, nuclear staining analysis in differentiating cultures revealed increased chromatin condensation and apoptosis in the Y218N at the late stage (Fig. 3d).
Transcriptional profile of maturating iPS cell cultures. (a) Quantitative RT-PCR transcriptional profile of control and mutant cell cultures at the three maturation stages. Bars represent the mean ± S.E.M. of 2–4 time points for each stage from at least 2 independent differentiations. Data are presented as mean ± standard error of the mean (S.E.M.). Differences between groups were considered statistically significant **** P < 0.001, *** P < 0.01 and ** P < 0.05. Bonferroni post hoc test. (b) Representative immunofluorescence microphotographs of PrPC, DCX and GFAP expression at the three differentiation stages. (c) Higher power image of GFAP positive cells at mid differentiation stage. (d). Quantification of apoptotic nuclei (% over total Hoechst). *** P < 0.01, Bonferroni post hoc test. Scale bars in b = 100 μm and c = 10 μm
Increased tau Phosphorylation in Y218N-Derived Neurons in Vitro
As indicated above, clinical and histopathological examination of the GSS patient carrying the Y218N PRNP mutation displayed relevant neurofibrillary degeneration with p-Tau deposits in several brain regions [17]. Therefore, we explored putative changes in Tau expression and phosphorylation in differentiating cultures (Fig. 4, Supplementary Figs. 3 and 4). As indicated above, MAP2 and MAPT mRNA levels decreased in Y218N-derived cultures compared to controls suggesting delayed neuronal differentiation (Fig. 3). This was corroborated analyzing the appearance of the two Tau-splicing forms (4R and 3R) in differentiated cultures (Fig. 4a). RT-qPCR analysis demonstrated a delayed appearance of the 4R Tau form compared to 3R Tau in Y218N-derived cultures (Fig. 4a). This was corroborated by the biochemical analysis of the acetylated form of Tau at lysine 280 (K280-(ac)) Tau during the differentiation (Fig. 4b, Supplementary Fig. 4). This acetylated form is associated with Tau 4R [42, 43]. K280-(ac) Tau levels were constant in control-derived cultures from 15 to 45 DIV. However, Y218N cultures showed increased levels of K280-(ac) Tau between 15 to 41 DIV, following changes of Tau 4R (Fig. 4b, Supplementary Fig. 4). In parallel, biochemical detection of p-Tau during differentiation demonstrated the increase in p-Tau (detected by AT8 and PHF1 antibodies) in Y218N-derived neurons compared to control without relevant changes in PrPC protein levels (Fig. 4b,c, Supplementary Fig. 3).
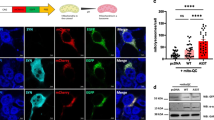
Delayed MAPT maturation and increased p-Tau in Y218N -derived neurons. (a) Histograms illustrating RT-qPCR results (mean ± S.E.M.) of Tau 3R/Total tau; Tau 4R/Total and Tau 3R/4R ratios in FHB1 and FH10 (Y218N) iPS cell cultures during differentiation at 15, 21 and 41 days in vitro. Asterisks in the right graph indicate P < 0.05, Bonferroni post hoc test; Mean Diff. -0.756; 95% confidence interval = −1.372 to - 0.1415). (b) Time course of p-Tau, PrPC and K280Tau-(ac) expression in FHB1 and FH10 (Y218N) at 15, 21 and 45 DIV. Actin was used as control loading protein. (c) Graph of the densitometric values of p-Tau levels of (b). Plots show mean ± S.E.M. of three different experiments. Note the increase in p-Tau between Y218N and control cells. (d) High power photomicrographs illustrating MAP2 (green), p-Tau (red) in FHB1 and FH10 (Y218N) neural cultures. A high magnification of a labelled cell is showed in (f). (e-f) Quantification of CTCF values derived from experiments in (d). Plots show mean ± S.E.M. of four different experiments. Asterisks in (e) indicate statistical differences between groups and controls. **** P < 0.001; Mann-Whitney U test. Scale bars in d = 50 μm and f = 10 μm
Next, we developed a CTCF analysis of p-Tau in identified MAP2-positive neurons (Fig. 4d,e). First we counted the total number of MAP2 and p-Tau-positive neurons in Y218N- and control-derived cultures. As suggested above with RT-qPCR, the total number of MAP2 and p-Tau double-positive cells was lower in Y218N-derived cultures (Fig. 4d,e). In addition, the relative percentage of double-labeled p-Tau/MAP2 neurons was also higher in Y218N-derived neurons. Changes in p-Tau level in differentiated neurons were also corroborated by the CTCF analysis of p-Tau in double-labeled p-Tau/MAP2-immunoreactive neurons (Control: 1566 ± 214.4; Y218N: 4357 ± 422.1; mean ± S.E.M. P < 0.0001, Mann-Whitney U test). Indeed, the ratio of p-Tau/MAP2 fluorescence was higher in Y218N-derived neurons compared to control-derived neurons (Control: 0.27 ± 0.02; Y218N: 0.680 ± 0.02; mean ± S.E.M. P < 0.0001, Mann-Whitney U = 2628) (Fig. 4e,f). Unfortunately, electron microscopy analyses failed to identify neurofibrillary tangle formation in Y218N differentiated neurons (not shown). Similar biochemical observations were also made using the direct cortical differentiation protocol (Supplementary Fig. 5 ). In conclusion, cultures derived from Y218N mutant iPS cells recapitulated in vitro several pathological features of the GSS patient, such as reactive astrocytosis, cell death and Tau hyperphosphorylation.
Impaired Mitochondria Movement in Y218N-Derived Neurons
The effects of Tau hyperphosphorylation in several epitopes on mitochondria movement have been demonstrated in Alzheimer’s disease [44, 28]. We checked the minimum and the mean velocity of identified mitochondria (Fig. 5). Results showed a decrease in both measurements in Y218N cultures compared with control (0.052 ± 0.014 (control, n = 111) vs 0.014 ± 0.006 (Y218N; n = 105); mean ± S.E.M., Min. velocity in μm/s; P = 0.0004; Mann-Whitney U value =4688. 0.310 ± 0.038 (control, n = 111) vs 0.1302 ± 0.014 (Y218N; n = 105); Mean ± S.E.M. Mean velocity in μm/s; P = 0.0484; Mann-Whitney U value =4921) (Fig. 5b).
FH10 (Y218N) cultures showed impaired mitochondria displacement. (a) Time-lapse fluorescence photomicrographs illustrating mitochondria movement in FHB1- (upper panels) and FH10 (Y218N)- (lower panels) derived neurons. The movement of two mitochondria (arrow and open arrow in (a) can be seen in the time lapse panels. (b) Plots illustrating the Minimum and Mean velocity values of tracked mitochondria in both types of cultures (see Methods for details). Notice the strong decreases in velocity in FH10 (Y218N)-derived cultures. Plots show mean ± S.E.M. of three different differentiation experiments. ***P < 0.01, ** P < 0.05. Mann Whitney U test. Scale bar: a = 2.5 μm
Infectivity Assays
In order to examine susceptibility to prion infection, Y218N- and control-differentiated cultures were exposed for 10 days to brain homogenates prepared from a sporadic CJD and from a GSS Y218N brain (Fig. 6a,b). Western blot analyses of the cultures showed the presence of PK-resistant PrP forms but only up to 2 weeks after removal of the inocula, indicating lack of prion infectivity (and propagation) in these conditions (Fig. 6c). A similar signal was observed in cultures infected with 263 K, a hamster scrapie prion strain which does not propagate to human cells (data not shown). Despite the absence of PK-resistant PrP in the cultures after two weeks, we observed some phenotypic changes that were more prominent in mutant neurons. In particular, we found a prominent redistribution of Tau signal with enhanced localization in the soma and proximal neurites in Y218N neurons exposed to either GSS or CJD inoculates was found (Fig. 6d,e).
Infectivity assay with brain inoculates. (a) Schematic representation of the inoculation protocol: infective brain homogenates were added at day 0 and day 3 and removed at day 10; cells were subsequently passaged several times to remove the inocula. (b) Inocula from the sources (10% of brain homogenates, see Methods for details) were processed to show PK-resistant PrP signal. GSS: human brain diagnosed of Y218N. CJD: human brain diagnosed of a sporadic CJD MM1. CJD samples were digested with 10–50 μg/ml of proteinase K (PK) and subjected to a standard biochemical analysis. GSS sample was treated as an atypical prion sample (see Methods). The samples were analyzed using the monoclonal antibody 3F4. MW: Molecular marker. (c) Representative examples of Western blot detection of PK-resistant PrP forms following inoculation with CJD and GSS brain samples. Note that PK-resistant PrP was only detected (when present) for the first 2 weeks after the infection. (d) Morphological analyses 2 months later revealed little effect of these inoculates in control neurons while mutant Y218N cultures (e) showed fewer neurons with marked cytoplasmic redistribution of Tau signal (b, f, j) and enhanced immunoreactivity for GFAP. Scale bars: 25 μm
Discussion
In the present study we have developed, for the first time, an iPS cell model of a familial human prionopathy. The donor GSS patient carrying the Y218N PRNP mutation showed relevant gliosis, cell death, massive deposits of PrP and neurofibrillary degeneration in different brain regions [17]. Surprisingly, the presence of the Y218N mutation in other family member belies atypical parkinsonism phenotype instead of neurofibrillary degeneration [32]. Indeed, in parallel with the classical signs of GSS-associated degeneration, other clinical presentations, such as Alzheimer’s-type, frontotemporal-like dementia, parkinsonism, and atypical psychiatric disorders, have all been reported (e.g., [45]). These different clinical manifestations have also been found in family members with the same PRNP mutation, attributed to the distinct abnormal isoforms of prion protein and polymorphisms at codon 129 [46, 47]. In fact, in this case the two GSS patients differ at the codon 129 polymorphism (129MV [32] and 129VV in [17]), which may contribute to clinical differences between cases.
We used two different well-characterized procedures to differentiate the iPS cells into neurons [34, 26]. The first was directed to obtaining forebrain cortical neurons [34] and the second one was directed to maintaining regulated developmental steps during neural development [26]. With both protocols we obtained similar results being able to determine that Y218N-derived cultures showed relevant GFAP reactivity, cell death, neuronal Tau redistribution, elevated p-Tau levels and changes in mitochondrial trafficking. Despite being unable to reproduce the spontaneous generation of PK-resistant forms or enabling prion propagation after inoculation in Y218N-iPS cell derived cultures, the differentiated neural cells recapitulated most of the pathological features observed in the GSS patient’s brain. In this way, these Y218N-derived cultures could be used as an in vitro platform for neurodegenerative studies in familial prionopathies with the aim of characterizing the role of particular PRNP mutations in comorbid taupathies and cell death.
Unfortunately, Y218N cells did not generate PrP spontaneously and they were unable to propagate human prions (CJD and Y218N GSS prions) in vitro. This was disappointing but certainly not fully unexpected given that it has never been possible to propagate infectivity in primary neurons with human prions. Furthermore there are no studies of Y218N PrP propagation in vivo in contrast to other human mutations: i) P102L GSS human prion in P101L mice [48], ii) A117V GSS-derived human prion inoculum in AV117 PRNP mice [49] and iii) P102L, A117V or F198S PRNP mutations in bank voles [50].
Concerning the absence of endogenous prion generation in Y218N-derived neurons we might hypothesize, in a simplistic manner, that current in vitro times are not long enough for the endogenous generation of human PrPres, considering the clinical onset and evolution of the GSS patient. However, we believe that the current scenario is not as simple, and that other, as yet unknown factors with key functions in protein misfolding and propagation may be absent from our cultures. In vitro prion propagation (of mostly mouse adapted strains) has been developed in neural and non-neural cell lines [51], primary neuronal cultures [52], cerebellar organotypic slices [53], and, with some controversy, in neurospheres (e.g., [54]). However, human prions have not been propagated in neuronal cultures to date. In fact, a single study of Ladogama et al. reported the transmission of human prions but using neuroblastoma cells [55]. In addition, the propagation of human strains was more successful when prions had been previously adapted to mice (e.g., M1000 [56]). Although endogenous expression levels could be relevant, we cannot rule out the participation of other non-neuronal cells (microglial cells) and inflammatory processes in protein misfolding and propagation [57] which did not fully develop in our iPS cell cultures, in contrast to other 3D organotypic approaches that could be assayed in future experiments [58].
In our study, the differentiation of Y218N-derived iPS cells was protracted. It is well known that appropriate temporal and transcriptional levels of PRNP are required for the correct differentiation of human embryonic stem cells [59] as well as other neural stem cells in vivo [60] and in vitro [61]. In fact, early attempts to ablate PRNP in mice using constitutive promoters and large PRNP mutations were not viable because PRNP expression starts around E7.5 in neural tissue [62]. Indeed, PrPC is involved in several neural and non-neural developmental functions and its absence either delays or interferes with cell proliferation and maturation [63–65]. Although a clear explanation of the physiological impact of the Y218N mutation in these processes remains elusive, the mutation might induces aberrant folding of the protein [66], which may impair neuronal differentiation. In fact, in our experiments, mutant neurons showed decreased mRNA levels of CALB, VGLUT1 and MAP2 compared with control cells. In addition, Y218N-derived neurons showed very low numbers of Ca2+ transients analyzed by Fluo4-AM (data not shown).
The deposition of hyperphosphorylated forms of Tau (p-Tau) has been described in familial and sporadic forms of prion diseases and in the brains of patients with variant CJD. Elevated levels of Tau (p-Tau and total Tau) have also been reported in the cerebrospinal fluid (CSF) of patients with sporadic CJD [67]. In addition, rodents infected with BSE [68], 263 K [69] and human CJD [70] derived inocula also showed elevated levels of p-Tau. Indeed, neurons in encephalopathy-affected brain regions displaying PrP aggregates showed relevant Tau redistribution with increased perinuclear location. This perinuclear Tau reorganization was observed in CJD- and GSS-treated Y218N-derived neurons in this study. In this matter, it is well known that aggregated prion peptides [71], as well as infectious prions [70], may modulate microtubule dynamics and stability, which may also in turn implicate Tau distribution. The increased presence and neuronal redistribution of Tau likely has a direct effect on the neuropathological process triggered by prion presence, because PrPC binding to Tau is probably disrupted by the mutations (at least for P102L PRNP [18]). In fact, if we consider that Y218N might alter natural PrPC functions associated with Tau, the cellular responses mediated by sCJD and GSS prions might be exacerbated in the presence of the Y218N PRNP mutation.
In conclusion, we report here the use of iPS cell-derived neurons to investigate the putative roles of the Y218N PRNP mutation in neural differentiation, Tau phosphorylation and cell death. This approach provides a powerful in vitro system for functional analysis of pathways regulating PRNP function in human cortical neurons, cellular mechanisms regulating tau phosphorylation in these models and for the identification and testing of candidate disease-modifying compounds.
References
Ironside JW (1998) Prion diseases in man. J Pathol 186(3):227–234. doi:10.1002/(SICI)1096-9896(1998110)186:3<227::AID-PATH174>3.0.CO;2-3
Schmidt C, Haik S, Satoh K, Rabano A, Martinez-Martin P, Roeber S, Brandel JP, Calero-Lara M et al (2012) Rapidly progressive Alzheimer's disease: A multicenter update. J Alzheimers Dis 30(4):751–756. doi:10.3233/JAD-2012-120007
Hodges JR, Davies R, Xuereb J, Kril J, Halliday G (2003) Survival in frontotemporal dementia. Neurology 61(3):349–354
Roberson ED, Hesse JH, Rose KD, Slama H, Johnson JK, Yaffe K, Forman MS, Miller CA et al (2005) Frontotemporal dementia progresses to death faster than Alzheimer disease. Neurology 65(5):719–725. doi:10.1212/01.wnl.0000173837.82820.9f
Greenfield JG, Graham DI, Lantos PL (2002) Greenfield's neuropathology, 7th edn. Arnold, London, New York
Liberski PP, Ironside JW (2004) An outline of the neuropathology of transmissible spongiform encephalopathies (prion diseases). Folia Neuropathol 42(Suppl B):39–58
Iwasaki Y, Mori K, Ito M, Nokura K, Tatsumi S, Mimuro M, Kitamoto T, Yoshida M (2014) Gerstmann-Straeussler-Scheinker disease with P102L prion protein gene mutation presenting with rapidly progressive clinical course. Clin Neuropathol 33(5):344–353. doi:10.5414/NP300733
Collins S, McLean CA, Masters CL (2001) Gerstmann-Straussler-Scheinker syndrome,fatal familial insomnia, and kuru: A review of these less common human transmissible spongiform encephalopathies. J Clin Neurosci 8(5):387–397. doi:10.1054/jocn.2001.0919
Piccardo P, Seiler C, Dlouhy SR, Young K, Farlow MR, Prelli F, Frangione B, Bugiani O et al (1996) Proteinase-K-resistant prion protein isoforms in Gerstmann-Straussler-Scheinker disease (Indiana kindred). J Neuropathol Exp Neurol 55(11):1157–1163
Ishizawa K, Komori T, Shimazu T, Yamamoto T, Kitamoto T, Shimazu K, Hirose T (2002) Hyperphosphorylated tau deposition parallels prion protein burden in a case of Gerstmann-Straussler-Scheinker syndrome P102L mutation complicated with dementia. Acta Neuropathol 104(4):342–350. doi:10.1007/s00401-002-0547-3
Yamazaki M, Oyanagi K, Mori O, Kitamura S, Ohyama M, Terashi A, Kitamoto T, Katayama Y (1999) Variant Gerstmann-Straussler syndrome with the P105L prion gene mutation: An unusual case with nigral degeneration and widespread neurofibrillary tangles. Acta Neuropathol 98(5):506–511
Tranchant C, Sergeant N, Wattez A, Mohr M, Warter JM, Delacourte A (1997) Neurofibrillary tangles in Gerstmann-Straussler-Scheinker syndrome with the A117V prion gene mutation. J Neurol Neurosurg Psychiatry 63(2):240–246
Simpson M, Johanssen V, Boyd A, Klug G, Masters CL, Li QX, Pamphlett R, McLean C et al (2013) Unusual clinical and molecular-pathological profile of gerstmann-Straussler-Scheinker disease associated with a novel PRNP mutation (V176G). JAMA Neurol 70(9):1180–1185. doi:10.1001/jamaneurol.2013.165
Ghetti B, Tagliavini F, Giaccone G, Bugiani O, Frangione B, Farlow MR, Dlouhy SR (1994) Familial Gerstmann-Straussler-Scheinker disease with neurofibrillary tangles. Mol Neurobiol 8(1):41–48. doi:10.1007/BF02778006
Hsiao K, Dlouhy SR, Farlow MR, Cass C, Da Costa M, Conneally PM, Hodes ME, Ghetti B et al (1992) Mutant prion proteins in Gerstmann-Straussler-Scheinker disease with neurofibrillary tangles. Nat Genet 1(1):68–71. doi:10.1038/ng0492-68
Woulfe J, Kertesz A, Frohn I, Bauer S, George-Hyslop PS, Bergeron C (2005) Gerstmann-Straussler-Scheinker disease with the Q217R mutation mimicking frontotemporal dementia. Acta Neuropathol 110(3):317–319. doi:10.1007/s00401-005-1054-0
Alzualde A, Indakoetxea B, Ferrer I, Moreno F, Barandiaran M, Gorostidi A, Estanga A, Ruiz I et al (2010) A novel PRNP Y218N mutation in Gerstmann-Straussler-Scheinker disease with neurofibrillary degeneration. J Neuropathol Exp Neurol 69(8):789–800. doi:10.1097/NEN.0b013e3181e85737
Wang XF, Dong CF, Zhang J, Wan YZ, Li F, Huang YX, Han L, Shan B et al (2008) Human tau protein forms complex with PrP and some GSS- and fCJD-related PrP mutants possess stronger binding activities with tau in vitro. Mol Cell Biochem 310(1–2):49–55. doi:10.1007/s11010-007-9664-6
Takahashi K, Yamanaka S (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126(4):663–676. doi:10.1016/j.cell.2006.07.024
Takahashi K, Okita K, Nakagawa M, Yamanaka S (2007) Induction of pluripotent stem cells from fibroblast cultures. Nat Protoc 2(12):3081–3089. doi:10.1038/nprot.2007.418
Shi Y, Inoue H, Wu JC, Yamanaka S (2017) Induced pluripotent stem cell technology: A decade of progress. Nat Rev Drug Discov 16(2):115–130. doi:10.1038/nrd.2016.245
Rao M (2013) iPSC crowdsourcing: A model for obtaining large panels of stem cell lines for screening. Cell Stem Cell 13(4):389–391. doi:10.1016/j.stem.2013.09.005
Dolmetsch R, Geschwind DH (2011) The human brain in a dish: The promise of iPSC-derived neurons. Cell 145(6):831–834. doi:10.1016/j.cell.2011.05.034
Muratore CR, Rice HC, Srikanth P, Callahan DG, Shin T, Benjamin LN, Walsh DM, Selkoe DJ et al (2014) The familial Alzheimer's disease APPV717I mutation alters APP processing and tau expression in iPSC-derived neurons. Hum Mol Genet 23(13):3523–3536. doi:10.1093/hmg/ddu064
Schondorf DC, Aureli M, McAllister FE, Hindley CJ, Mayer F, Schmid B, Sardi SP, Valsecchi M et al (2014) iPSC-derived neurons from GBA1-associated Parkinson's disease patients show autophagic defects and impaired calcium homeostasis. Nat Commun 5:4028. doi:10.1038/ncomms5028
Sanchez-Danes A, Richaud-Patin Y, Carballo-Carbajal I, Jimenez-Delgado S, Caig C, Mora S, Di Guglielmo C, Ezquerra M et al (2012) Disease-specific phenotypes in dopamine neurons from human iPS-based models of genetic and sporadic Parkinson's disease. EMBO Mol Med 4(5):380–395. doi:10.1002/emmm.201200215
Jeon I, Lee N, Li JY, Park IH, Park KS, Moon J, Shim SH, Choi C et al (2012) Neuronal properties, in vivo effects, and pathology of a Huntington's disease patient-derived induced pluripotent stem cells. Stem Cells 30(9):2054–2062. doi:10.1002/stem.1135
Iovino M, Agathou S, Gonzalez-Rueda A, Del Castillo V-HM, Borroni B, Alberici A, Lynch T, O'Dowd S et al (2015) Early maturation and distinct tau pathology in induced pluripotent stem cell-derived neurons from patients with MAPT mutations. Brain 138(Pt 11):3345–3359. doi:10.1093/brain/awv222
Zhang K, Donnelly CJ, Haeusler AR, Grima JC, Machamer JB, Steinwald P, Daley EL, Miller SJ et al (2015) The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature 525(7567):56–61. doi:10.1038/nature14973
Zhang Y, Pak C, Han Y, Ahlenius H, Zhang Z, Chanda S, Marro S, Patzke C et al (2013) Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron 78(5):785–798. doi:10.1016/j.neuron.2013.05.029
Canals I, Soriano J, Orlandi JG, Torrent R, Richaud-Patin Y, Jimenez-Delgado S, Merlin S, Follenzi A et al (2015) Activity and high-order effective connectivity alterations in Sanfilippo C patient-specific neuronal networks. Stem Cell Reports 5(4):546–557. doi:10.1016/j.stemcr.2015.08.016
Ribosa-Nogue R, Pagonabarraga J, Gomez-Anson B, Granell-Moreno E, Sanchez-Valle R, Kulisevsky J (2015) Gerstmann-Straussler-Scheinker Disease Presenting with Atypical Parkinsonism, but Typical Magnetic Resonance Imaging Findings of Prion Disease. Movement Disorders:93–95.
Raya A, Rodriguez-Piza I, Guenechea G, Vassena R, Navarro S, Barrero MJ, Consiglio A, Castella M et al (2009) Disease-corrected haematopoietic progenitors from Fanconi anaemia induced pluripotent stem cells. Nature 460(7251):53–59. doi:10.1038/nature08129
Espuny-Camacho I, Michelsen KA, Gall D, Linaro D, Hasche A, Bonnefont J, Bali C, Orduz D et al (2013) Pyramidal neurons derived from human pluripotent stem cells integrate efficiently into mouse brain circuits in vivo. Neuron 77(3):440–456. doi:10.1016/j.neuron.2012.12.011
Gaspard N, Bouschet T, Herpoel A, Naeije G, van den Ameele J, Vanderhaeghen P (2009) Generation of cortical neurons from mouse embryonic stem cells. Nat Protoc 4(10):1454–1463. doi:10.1038/nprot.2009.157
Aguila JC, Blak A, van Arensbergen J, Sousa A, Vazquez N, Aduriz A, Gayosso M, Lopez Mato MP et al (2014) Selection based on FOXA2 expression is not sufficient to enrich for dopamine neurons from human pluripotent stem cells. Stem Cells Transl Med 3(9):1032–1042. doi:10.5966/sctm.2014-0011
Cho MS, Hwang DY, Kim DW (2008) Efficient derivation of functional dopaminergic neurons from human embryonic stem cells on a large scale. Nat Protoc 3(12):1888–1894. doi:10.1038/nprot.2008.188
McCloy RA, Rogers S, Caldon CE, Lorca T, Castro A, Burgess A (2014) Partial inhibition of Cdk1 in G 2 phase overrides the SAC and decouples mitotic events. Cell Cycle 13(9):1400–1412. doi:10.4161/cc.28401
Oelschlegel AM, Geissen M, Lenk M, Riebe R, Angermann M, Schatzl H, Groschup MH (2015) A bovine cell line that can be infected by natural sheep scrapie prions. PLoS One 10(1):e0117154. doi:10.1371/journal.pone.0117154
Pirisinu L, Nonno R, Esposito E, Benestad SL, Gambetti P, Agrimi U, Zou WQ (2013) Small ruminant nor98 prions share biochemical features with human gerstmann-straussler-scheinker disease and variably protease-sensitive prionopathy. PLoS One 8(6):e66405. doi:10.1371/journal.pone.0066405
Bendheim PE, Brown HR, Rudelli RD, Scala LJ, Goller NL, Wen GY, Kascsak RJ, Cashman NR et al (1992) Nearly ubiquitous tissue distribution of the scrapie agent precursor protein. Neurology 42(1):149–156
Grinberg LT, Wang X, Wang C, Sohn PD, Theofilas P, Sidhu M, Arevalo JB, Heinsen H et al (2013) Argyrophilic grain disease differs from other tauopathies by lacking tau acetylation. Acta Neuropathol 125(4):581–593. doi:10.1007/s00401-013-1080-2
Cohen TJ, Guo JL, Hurtado DE, Kwong LK, Mills IP, Trojanowski JQ, Lee VM (2011) The acetylation of tau inhibits its function and promotes pathological tau aggregation. Nat Commun 2:252. doi:10.1038/ncomms1255
Shahpasand K, Uemura I, Saito T, Asano T, Hata K, Shibata K, Toyoshima Y, Hasegawa M et al (2012) Regulation of mitochondrial transport and inter-microtubule spacing by tau phosphorylation at the sites hyperphosphorylated in Alzheimer's disease. J Neurosci 32(7):2430–2441. doi:10.1523/JNEUROSCI.5927-11.2012
Webb TE, Poulter M, Beck J, Uphill J, Adamson G, Campbell T, Linehan J, Powell C et al (2008) Phenotypic heterogeneity and genetic modification of P102L inherited prion disease in an international series. Brain 131(Pt 10):2632–2646. doi:10.1093/brain/awn202
Furukawa H, Doh-ura K, Kikuchi H, Tateishi J, Iwaki T (1998) A comparative study of abnormal prion protein isoforms between Gerstmann-Straussler-Scheinker syndrome and Creutzfeldt-Jakob disease. J Neurol Sci 158(1):71–75
Parchi P, Chen SG, Brown P, Zou W, Capellari S, Budka H, Hainfellner J, Reyes PF et al (1998) Different patterns of truncated prion protein fragments correlate with distinct phenotypes in P102L Gerstmann-Straussler-Scheinker disease. Proc Natl Acad Sci U S A 95(14):8322–8327
Asante EA, Grimshaw A, Smidak M, Jakubcova T, Tomlinson A, Jeelani A, Hamdan S, Powell C et al (2015) Transmission properties of human PrP 102L prions challenge the relevance of mouse models of GSS. PLoS Pathog 11(7):e1004953. doi:10.1371/journal.ppat.1004953
Asante EA, Linehan JM, Smidak M, Tomlinson A, Grimshaw A, Jeelani A, Jakubcova T, Hamdan S et al (2013) Inherited prion disease A117V is not simply a proteinopathy but produces prions transmissible to transgenic mice expressing homologous prion protein. PLoS Pathog 9(9):e1003643. doi:10.1371/journal.ppat.1003643
Pirisinu L, Di Bari MA, D'Agostino C, Marcon S, Riccardi G, Poleggi A, Cohen ML, Appleby BS et al (2016) Gerstmann-Straussler-Scheinker disease subtypes efficiently transmit in bank voles as genuine prion diseases. Scientific reports 6:20443. doi:10.1038/srep20443
Klohn PC, Stoltze L, Flechsig E, Enari M, Weissmann C (2003) A quantitative, highly sensitive cell-based infectivity assay for mouse scrapie prions. Proc Natl Acad Sci U S A 100(20):11666–11671. doi:10.1073/pnas.1834432100
Cronier S, Laude H, Peyrin JM (2004) Prions can infect primary cultured neurons and astrocytes and promote neuronal cell death. Proc Natl Acad Sci U S A 101(33):12271–12276. doi:10.1073/pnas.0402725101
Falsig J, Aguzzi A (2008) The prion organotypic slice culture assay--POSCA. Nat Protoc 3(4):555–562. doi:10.1038/nprot.2008.13
Giri RK, Young R, Pitstick R, DeArmond SJ, Prusiner SB, Carlson GA (2006) Prion infection of mouse neurospheres. Proc Natl Acad Sci U S A 103(10):3875–3880. doi:10.1073/pnas.0510902103
Ladogana A, Liu Q, Xi YG, Pocchiari M (1995) Proteinase-resistant protein in human neuroblastoma cells infected with brain material from Creutzfeldt-Jakob patient. Lancet 345(8949):594–595
Lewis V, Hill AF, Haigh CL, Klug GM, Masters CL, Lawson VA, Collins SJ (2009) Increased proportions of C1 truncated prion protein protect against cellular M1000 prion infection. J Neuropathol Exp Neurol 68(10):1125–1135. doi:10.1097/NEN.0b013e3181b96981
Heikenwalder M, Zeller N, Seeger H, Prinz M, Klohn PC, Schwarz P, Ruddle NH, Weissmann C et al (2005) Chronic lymphocytic inflammation specifies the organ tropism of prions. Science 307(5712):1107–1110. doi:10.1126/science.1106460
Falsig J, Julius C, Margalith I, Schwarz P, Heppner FL, Aguzzi A (2008) A versatile prion replication assay in organotypic brain slices. Nat Neurosci 11(1):109–117. doi:10.1038/nn2028
Lee YJ, Baskakov IV (2014) The cellular form of the prion protein guides the differentiation of human embryonic stem cells into neuron-, oligodendrocyte-, and astrocyte-committed lineages. Prion 8(3):266–275. doi:10.4161/pri.32079
Steele AD, Emsley JG, Ozdinler PH, Lindquist S, Macklis JD (2006) Prion protein (PrPc) positively regulates neural precursor proliferation during developmental and adult mammalian neurogenesis. Proc Natl Acad Sci U S A 103(9):3416–3421. doi:10.1073/pnas.0511290103
Santos TG, Silva IR, Costa-Silva B, Lepique AP, Martins VR, Lopes MH (2011) Enhanced neural progenitor/stem cells self-renewal via the interaction of stress-inducible protein 1 with the prion protein. Stem Cells 29(7):1126–1136. doi:10.1002/stem.664
Tremblay P, Bouzamondo-Bernstein E, Heinrich C, Prusiner SB, DeArmond SJ (2007) Developmental expression of PrP in the post-implantation embryo. Brain Res 1139:60–67. doi:10.1016/j.brainres.2006.12.055
Del Rio JA, Gavin R (2016) Functions of the cellular prion protein, the end of Moore's law, and Ockham's razor theory. Prion 10(1):25–40. doi:10.1080/19336896.2015.1126038
Linden R, Martins VR, Prado MA, Cammarota M, Izquierdo I, Brentani RR (2008) Physiology of the prion protein. Physiol Rev 88(2):673–728. doi:10.1152/physrev.00007.2007
Llorens F, Carulla P, Villa A, Torres JM, Fortes P, Ferrer I, del Rio JA (2013) PrP(C) regulates epidermal growth factor receptor function and cell shape dynamics in Neuro2a cells. J Neurochem 127(1):124–138. doi:10.1111/jnc.12283
Cheng CJ, Daggett V (2014) Different misfolding mechanisms converge on common conformational changes: Human prion protein pathogenic mutants Y218N and E196K. Prion 8(1):125–135
Otto M, Wiltfang J, Tumani H, Zerr I, Lantsch M, Kornhuber J, Weber T, Kretzschmar HA et al (1997) Elevated levels of tau-protein in cerebrospinal fluid of patients with Creutzfeldt-Jakob disease. Neurosci Lett 225(3):210–212
Bautista MJ, Gutierrez J, Salguero FJ, Fernandez de Marco MM, Romero-Trevejo JL, Gomez-Villamandos JC (2006) BSE infection in bovine PrP transgenic mice leads to hyperphosphorylation of tau-protein. Vet Microbiol 115(4):293–301. doi:10.1016/j.vetmic.2006.02.017
Wang GR, Shi S, Gao C, Zhang BY, Tian C, Dong CF, Zhou RM, Li XL et al (2010) Changes of tau profiles in brains of the hamsters infected with scrapie strains 263 K or 139 a possibly associated with the alteration of phosphate kinases. BMC Infect Dis 10:86. doi:10.1186/1471-2334-10-86
Lawson VA, Klemm HM, Welton JM, Masters CL, Crouch P, Cappai R, Ciccotosto GD (2011) Gene knockout of tau expression does not contribute to the pathogenesis of prion disease. J Neuropathol Exp Neurol 70(11):1036–1045. doi:10.1097/NEN.0b013e318235b471
Brown DR (2000) Altered toxicity of the prion protein peptide PrP106-126 carrying the ala(117)-->Val mutation. Biochem J 346(Pt 3):785–791
Acknowledgements
The authors thank Tom Yohannan for editorial advice and M. Segura-Feliu for technical assistance. We thank members of the Del Río, Sánchez-Pernaute, Castilla, Raya, Consiglio, López de Munain and Ferrer groups for stimulating discussions and ideas. We also thank to Lidia Bardia of the Advanced Confocal Microscopy service of the IRB-PCB for helping us with mitochondria tracking experiments. This research was supported by grants from the Spanish Ministry of Economy, Industry and Competitiveness (MINECO) (BFU2015-67777-R), the Spanish prion network (Prionet Spain, AGL2015-71764-REDT), the Generalitat de Catalunya (SGR2014-1218), CIBERNED (PI2014/02-4 (Rapid dementias) and PI2016/02 (MFDEND)), La Caixa Obra Social Foundation, La Marató de TV3 to J.A.D.R.. R.S.P was funded by the Joint Program in Neurodegenerative Diseases (DAMNDPATHS, AC14/00021). J.C. was funded by MINECO (AGL2015-65046-C2-1-R) and EiTB Maratoia (BioEF): BIO12/AL/004. A.R. was supported by MINECO (SAF2015-69706-R), Instituto de Salud Carlos III/FEDER (TerCel RD16/0011/0024, PIE14/00061), AGAUR (2014-SGR-1460), and Fundació La Marató de TV3 (201534-30) and CERCA Programme/Generalitat de Catalunya. I.F. was funded was supported by grants from CIBERNED (PI2014/02-4 Rapid dementias) and Fondo de Investigaciones Sanitarias (Instituto de Salud Carlos III), and co-funded by FEDER funds/European Regional Development Fund (ERDF) – a way to build Europe PIE14/00034 and PI14/00757. A.C. was also supported by BFU2013-49157-P and RETICTerCel grants from MINECO and the European Research Council (ERC) 2012-StG (311736- PD-HUMMODEL). A.L.M. was funded by grants from Fondo de Investigaciones Sanitarias (Instituto de Salud Carlos III (PI14/00436)), and the Joint Program in Neurodegenerative Diseases (DEMTEST). A.M-A. was supported by a fellowship from the Fundación Tatiana Pérez de Guzmán el Bueno. L.M.G. was supported by a CONACYT fellowship (CVU 357631, Mexico). The authors declare no conflicts of interest.
Author Contribution
A.M-A.; L.M.G; Y.R-P. performed most of the experiments. Y.R-P.; A.D.D.; A.C.; A.R. generated the iPS cell lines and developed the second protocol of neuronal differentiation and their characterization with C.V.; R.G.; A,H.; I.F. and A.M-A.; A.S. L.M.G and R. S-P. developed the first protocol of iPS differentiation and their characterization with RL-M. L.M.G and N.F.B performed the infectivity assays and the analysis of results with J.C. and R. S-P. A.L.M.; A.R.; J.C.; R.S-P.; J.A.D.R. designed and supervised the experiments and wrote the manuscript. All authors reviewed the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Andreu Matamoros-Angles, Lucía Mayela Gayosso, and Yvonne Richaud-Patin contribute equally to this study.
Electronic supplementary material
Supplementary Table 1
(DOCX 14 kb)
Supplementary Table 2
(DOCX 15.3 kb)
Supplementary Table 3
(DOCX 13.6 kb)
Supplementary Fig. 1
Full size uncropped blots corresponding to Fig. 2g, IPS (asterisk in a) and SNM (asterisk in b). (JPEG 4.13 mb)
Supplementary Fig. 2
High ECL exposure (15 min) showing the determination of PK-resistant PrPC levels illustrated in Fig. 2h. Uncropped film. (JPEG 621 kb)
Supplementary Fig. 4
Full size uncropped blots corresponding to Fig. 4b (lower panels). (JPEG 1.15 mb)
Supplementary Fig. 5
Determination of p-Tau (AT8 antibody) levels in FHB1 and FH10 (Y218N)-derived cultures at several DIV of differentiation (procedure 1, see Methods). p-Tau probed membranes were immunoblotted using antibodies against GADPH for standardization. (JPEG 488 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Matamoros-Angles, A., Gayosso, L.M., Richaud-Patin, Y. et al. iPS Cell Cultures from a Gerstmann-Sträussler-Scheinker Patient with the Y218N PRNP Mutation Recapitulate tau Pathology. Mol Neurobiol 55, 3033–3048 (2018). https://doi.org/10.1007/s12035-017-0506-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-017-0506-6