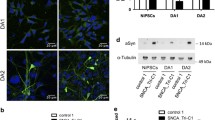
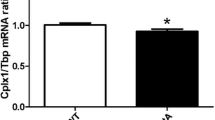
Abstract
Alpha-synuclein is an abundant neuronal protein which has been associated with physiological processes like synaptic function, neurogenesis, and neuronal differentiation but also with pathological neurodegeneration. Indeed, alpha-synuclein (snca) is one of the major genes implicated in Parkinson’s disease (PD). However, little is known about the regulation of alpha-synuclein expression. Unveiling the mechanisms that control its regulation is of high importance, as it will enable to further investigate and comprehend the physiological role of alpha-synuclein as well as its potential contribution in the aetiology of PD. Previously, we have shown that the protein TRIM32 regulates fate specification of neural stem cells. Here, we investigated the impact of TRIM32 on snca expression regulation in vitro and in vivo in neural stem cells and neurons. We demonstrated that TRIM32 is positively influencing snca expression in a neuronal cell line, while the absence of TRIM32 is causing deregulated levels of snca transcripts. Finally, we provided evidence that TRIM32 binds to the promoter region of snca, suggesting a novel mechanism of its transcriptional regulation. On the one hand, the presented data link the PD-associated gene alpha-synuclein to the neuronal cell fate determinant TRIM32 and thereby support the concept that PD is a neurodevelopmental disorder. On the other hand, they imply that defects in olfactory bulb adult neurogenesis might contribute to early PD-associated non-motor symptoms like hyposmia.





Similar content being viewed by others
References
de Rijk MC, Launer LJ, Berger K, Breteler MM, Dartigues JF, Baldereschi M, Fratiglioni L, Lobo A et al (2000) Prevalence of Parkinson’s disease in Europe: a collaborative study of population-based cohorts. Neurologic Diseases in the Elderly Research Group. Neurology 54(11 Suppl 5):S21–3
de Lau LM, Breteler MM (2006) Epidemiology of Parkinson’s disease. Lancet Neurol 5(6):525–35. doi:10.1016/S1474-4422(06)70471-9
Van Den Eeden SK, Tanner CM, Bernstein AL, Fross RD, Leimpeter A, Bloch DA, Nelson LM (2003) Incidence of Parkinson’s disease: variation by age, gender, and race/ethnicity. Am J Epidemiol 157(11):1015–22
Shulman JM, De Jager PL, Feany MB (2011) Parkinson’s disease: genetics and pathogenesis. Annu Rev Pathol 6:193–222. doi:10.1146/annurev-pathol-011110-130242
Vekrellis K, Rideout HJ, Stefanis L (2004) Neurobiology of alpha-synuclein. Mol Neurobiol 30(1):1–21. doi:10.1385/MN:30:1:001
Braak H, Del Tredici K, Rub U, de Vos RA, Jansen Steur EN, Braak E (2003) Staging of brain pathology related to sporadic Parkinson’s disease. Neurobiol Aging 24(2):197–211
Gasser T (2009) Molecular pathogenesis of Parkinson disease: insights from genetic studies. Expert Rev Mol Med 11:e22. doi:10.1017/S1462399409001148
Nuytemans K, Theuns J, Cruts M, Van Broeckhoven C (2010) Genetic etiology of Parkinson disease associated with mutations in the SNCA, PARK2, PINK1, PARK7, and LRRK2 genes: a mutation update. Hum Mutat 31(7):763–80. doi:10.1002/humu.21277
Sundal C, Fujioka S, Uitti RJ, Wszolek ZK (2012) Autosomal dominant Parkinson’s disease. Parkinsonism Relat Disord 18(Suppl 1):S7–10. doi:10.1016/S1353-8020(11)70005-0
Golbe LI, Di Iorio G, Sanges G, Lazzarini AM, La Sala S, Bonavita V, Duvoisin RC (1996) Clinical genetic analysis of Parkinson’s disease in the Contursi kindred. Ann Neurol 40(5):767–75. doi:10.1002/ana.410400513
Pearce RK, Hawkes CH, Daniel SE (1995) The anterior olfactory nucleus in Parkinson’s disease. Mov Disord 10(3):283–7. doi:10.1002/mds.870100309
Del Tredici K, Rub U, De Vos RA, Bohl JR, Braak H (2002) Where does parkinson disease pathology begin in the brain? J Neuropathol Exp Neurol 61(5):413–26
Tani M, Hayakawa H, Yasuda T, Nihira T, Hattori N, Mizuno Y, Mochizuki H (2010) Ectopic expression of alpha-synuclein affects the migration of neural stem cells in mouse subventricular zone. J Neurochem 115(4):854–63. doi:10.1111/j.1471-4159.2010.06727.x
Crews L, Mizuno H, Desplats P, Rockenstein E, Adame A, Patrick C, Winner B, Winkler J et al (2008) Alpha-synuclein alters Notch-1 expression and neurogenesis in mouse embryonic stem cells and in the hippocampus of transgenic mice. J Neurosci 28(16):4250–60. doi:10.1523/JNEUROSCI.0066-08.2008
Desplats P, Spencer B, Crews L, Pathel P, Morvinski-Friedmann D, Kosberg K, Roberts S, Patrick C et al (2012) Alpha-synuclein induces alterations in adult neurogenesis in Parkinson disease models via p53-mediated repression of Notch1. J Biol Chem 287(38):31691–702. doi:10.1074/jbc.M112.354522
Winner B, Lie DC, Rockenstein E, Aigner R, Aigner L, Masliah E, Kuhn HG, Winkler J (2004) Human wild-type alpha-synuclein impairs neurogenesis. J Neuropathol Exp Neurol 63(11):1155–66
Alvarez-Buylla A, Garcia-Verdugo JM (2002) Neurogenesis in adult subventricular zone. J Neurosci 22(3):629–34
Lazarini F, Lledo PM (2011) Is adult neurogenesis essential for olfaction? Trends Neurosci 34(1):20–30. doi:10.1016/j.tins.2010.09.006
Feierstein CE, Lazarini F, Wagner S, Gabellec MM, de Chaumont F, Olivo-Marin JC, Boussin FD, Lledo PM et al (2010) Disruption of adult neurogenesis in the olfactory bulb affects social interaction but not maternal behavior. Front Behav Neurosci 4:176. doi:10.3389/fnbeh.2010.00176
Hillje A-L, Beckmann E, Pavlou M, Jaeger C, Pacheco MP, Sauter T, Hiller K, Schwamborn JC, et al. (2015) The neural stem cell fate determinant TRIM32 regulates complex behavioral traits. Front Cell Neurosc 9. doi:10.3389/fncel.2015.00075
Marxreiter F, Regensburger M, Winkler J (2013) Adult neurogenesis in Parkinson’s disease. Cell Mol Life Sci 70(3):459–73. doi:10.1007/s00018-012-1062-x
Gonzalez-Cano L, Hillje AL, Fuertes-Alvarez S, Marques MM, Blanch A, Ian RW, Irwin MS, Schwamborn JC et al (2013) Regulatory feedback loop between TP73 and TRIM32. Cell Death Dis 4:e704. doi:10.1038/cddis.2013.224
Schwamborn JC, Berezikov E, Knoblich JA (2009) The TRIM-NHL protein TRIM32 activates microRNAs and prevents self-renewal in mouse neural progenitors. Cell 136(5):913–25. doi:10.1016/j.cell.2008.12.024
Nicklas S, Okawa S, Hillje AL, Gonzalez-Cano L, Del Sol A, Schwamborn JC (2015) The RNA helicase DDX6 regulates cell-fate specification in neural stem cells via miRNAs. Nucleic Acids Res 43(5):2638–54. doi:10.1093/nar/gkv138
Hillje AL, Worlitzer MM, Palm T, Schwamborn JC (2011) Neural stem cells maintain their stemness through protein kinase C zeta-mediated inhibition of TRIM32. Stem Cells 29(9):1437–47. doi:10.1002/stem.687
Hillje AL, Pavlou MA, Beckmann E, Worlitzer MM, Bahnassawy L, Lewejohann L, Palm T, Schwamborn JC (2013) TRIM32-dependent transcription in adult neural progenitor cells regulates neuronal differentiation. Cell Death Dis 4:e976. doi:10.1038/cddis.2013.487
Clough RL, Dermentzaki G, Stefanis L (2009) Functional dissection of the alpha-synuclein promoter: transcriptional regulation by ZSCAN21 and ZNF219. J Neurochem 110(5):1479–90. doi:10.1111/j.1471-4159.2009.06250.x
Scherzer CR, Grass JA, Liao Z, Pepivani I, Zheng B, Eklund AC, Ney PA, Ng J et al (2008) GATA transcription factors directly regulate the Parkinson’s disease-linked gene alpha-synuclein. Proc Natl Acad Sci U S A 105(31):10907–12. doi:10.1073/pnas.0802437105
Yang YX, Latchman DS (2008) Nurr1 transcriptionally regulates the expression of alpha-synuclein. Neuroreport 19(8):867–71. doi:10.1097/WNR.0b013e3282ffda48
Hillje AL, Worlitzer MM, Palm T, Schwamborn JC (2011) Neural stem cells maintain their stemness through protein kinase C zeta-mediated inhibition of TRIM32. Stem Cells 29((9):1437–47. doi:10.1002/stem.687;10.1002/stem.687, Dayton, Ohio
Szpara ML, Vranizan K, Tai YC, Goodman CS, Speed TP, Ngai J (2007) Analysis of gene expression during neurite outgrowth and regeneration. BMC Neurosci 8:100. doi:10.1186/1471-2202-8-100
Ljung L (1998) System identification. In: Procházka A, Uhlíř J, Rayner PWJ, Kingsbury NG (eds) Signal analysis and prediction. Applied and numerical harmonic analysis. Birkhäuser, Boston, pp 163–73. doi:10.1007/978-1-4612-1768-8_11
Chiba-Falek O, Nussbaum RL (2001) Effect of allelic variation at the NACP-Rep1 repeat upstream of the alpha-synuclein gene (SNCA) on transcription in a cell culture luciferase reporter system. Hum Mol Genet 10(26):3101–9
Hegde ML, Jagannatha Rao KS (2003) Challenges and complexities of alpha-synuclein toxicity: new postulates in unfolding the mystery associated with Parkinson’s disease. Arch Biochem Biophys 418(2):169–78
Satoh JI, Kuroda Y (2001) Alpha-synuclein expression is up-regulated in NTera2 cells during neuronal differentiation but unaffected by exposure to cytokines and neurotrophic factors. Parkinsonism Relat Disord 8(1):7–17
Specht CG, Schoepfer R (2001) Deletion of the alpha-synuclein locus in a subpopulation of C57BL/6J inbred mice. BMC Neurosci 2:11
Frosk P, Weiler T, Nylen E, Sudha T, Greenberg CR, Morgan K, Fujiwara TM, Wrogemann K (2002) Limb-girdle muscular dystrophy type 2H associated with mutation in TRIM32, a putative E3-ubiquitin-ligase gene. Am J Hum Genet 70(3):663–72. doi:10.1086/339083
Chiang AP, Beck JS, Yen HJ, Tayeh MK, Scheetz TE, Swiderski RE, Nishimura DY, Braun TA et al (2006) Homozygosity mapping with SNP arrays identifies TRIM32, an E3 ubiquitin ligase, as a Bardet-Biedl syndrome gene (BBS11). Proc Natl Acad Sci U S A 103(16):6287–92. doi:10.1073/pnas.0600158103
Kano S, Miyajima N, Fukuda S, Hatakeyama S (2008) Tripartite motif protein 32 facilitates cell growth and migration via degradation of Abl-interactor 2. Cancer Res 68(14):5572–80. doi:10.1158/0008-5472.CAN-07-6231
Albor A, Kulesz-Martin M (2007) Novel initiation genes in squamous cell carcinomagenesis: a role for substrate-specific ubiquitylation in the control of cell survival. Mol Carcinog 46(8):585–90. doi:10.1002/mc.20344
Lionel AC, Tammimies K, Vaags AK, Rosenfeld JA, Ahn JW, Merico D, Noor A, Runke CK et al (2014) Disruption of the ASTN2/TRIM32 locus at 9q33.1 is a risk factor in males for autism spectrum disorders, ADHD and other neurodevelopmental phenotypes. Hum Mol Genet 23(10):2752–68. doi:10.1093/hmg/ddt669
Lo-Castro A, Curatolo P (2014) Epilepsy associated with autism and attention deficit hyperactivity disorder: is there a genetic link? Brain Dev 36(3):185–93. doi:10.1016/j.braindev.2013.04.013
Ruan CS, Wang SF, Shen YJ, Guo Y, Yang CR, Zhou FH, Tan LT, Zhou L et al (2014) Deletion of TRIM32 protects mice from anxiety- and depression-like behaviors under mild stress. Eur J Neurosci 40(4):2680–90. doi:10.1111/ejn.12618
Horn EJ, Albor A, Liu Y, El-Hizawi S, Vanderbeek GE, Babcock M, Bowden GT, Hennings H et al (2004) RING protein Trim32 associated with skin carcinogenesis has anti-apoptotic and E3-ubiquitin ligase properties. Carcinogenesis 25(2):157–67. doi:10.1093/carcin/bgh003
Albor A, El-Hizawi S, Horn EJ, Laederich M, Frosk P, Wrogemann K, Kulesz-Martin M (2006) The interaction of Piasy with Trim32, an E3-ubiquitin ligase mutated in limb-girdle muscular dystrophy type 2H, promotes Piasy degradation and regulates UVB-induced keratinocyte apoptosis through NFkappaB. J Biol Chem 281(35):25850–66. doi:10.1074/jbc.M601655200
Ryu YS, Lee Y, Lee KW, Hwang CY, Maeng JS, Kim JH, Seo YS, You KH et al (2011) TRIM32 protein sensitizes cells to tumor necrosis factor (TNFalpha)-induced apoptosis via its RING domain-dependent E3 ligase activity against X-linked inhibitor of apoptosis (XIAP). J Biol Chem 286(29):25729–38. doi:10.1074/jbc.M111.241893
Liu J, Zhang C, Wang XL, Ly P, Belyi V, Xu-Monette ZY, Young KH, Hu W et al (2014) E3 ubiquitin ligase TRIM32 negatively regulates tumor suppressor p53 to promote tumorigenesis. Cell Death Differ 21(11):1792–804. doi:10.1038/cdd.2014.121
Zhang S, Xiao Q, Le W (2015) olfactory dysfunction and neurotransmitter disturbance in olfactory bulb of transgenic mice expressing human A53T mutant alpha-synuclein. PLoS One 10(3):e0119928. doi:10.1371/journal.pone.0119928
Mahlknecht P, Iranzo A, Hogl B, Frauscher B, Muller C, Santamaria J, Tolosa E et al (2015) Olfactory dysfunction predicts early transition to a Lewy body disease in idiopathic RBD. Neurology 84(7):654–8. doi:10.1212/WNL.0000000000001265
Jowaed A, Schmitt I, Kaut O, Wullner U (2010) Methylation regulates alpha-synuclein expression and is decreased in Parkinson’s disease patients’ brains. J Neurosci 30(18):6355–9. doi:10.1523/JNEUROSCI.6119-09.2010
Matsumoto L, Takuma H, Tamaoka A, Kurisaki H, Date H, Tsuji S, Iwata A (2010) CpG demethylation enhances alpha-synuclein expression and affects the pathogenesis of Parkinson’s disease. PLoS One 5:e15522–e15522. doi:10.1371/journal.pone.001552210.1371/journal.pone.0015522
Pihlstrom L, Berge V, Rengmark A, Toft M (2015) Parkinson’s disease correlates with promoter methylation in the alpha-synuclein gene. Mov Disord 30(4):577–80. doi:10.1002/mds.26073
Tan YY, Wu L, Zhao ZB, Wang Y, Xiao Q, Liu J, Wang G, Ma JF, Chen SD (2014) Methylation of alpha-synuclein and leucine-rich repeat kinase 2 in leukocyte DNA of Parkinson’s disease patients. Parkinsonism Relat Disord 20:308–313. doi:10.1016/j.parkreldis.2013.12.002
Chiba-Falek O, Touchman JW, Nussbaum RL (2003) Functional analysis of intra-allelic variation at NACP-Rep1 in the alpha-synuclein gene. Hum Genet 113(5):426–31. doi:10.1007/s00439-003-1002-9
Cronin KD, Ge D, Manninger P, Linnertz C, Rossoshek A, Orrison BM, Bernard DJ, El-Agnaf OM et al (2009) Expansion of the Parkinson disease-associated SNCA-Rep1 allele upregulates human alpha-synuclein in transgenic mouse brain. Hum Mol Genet 18(17):3274–85. doi:10.1093/hmg/ddp265
Hasegawa T, Matsuzaki M, Takeda A, Kikuchi A, Akita H, Perry G, Smith MA, Itoyama Y (2004) Accelerated alpha-synuclein aggregation after differentiation of SH-SY5Y neuroblastoma cells. Brain Res 1013(1):51–9. doi:10.1016/j.brainres.2004.04.018
Le Grand JN, Gonzalez-Cano L, Pavlou MA, Schwamborn JC (2015) Neural stem cells in Parkinson’s disease: a role for neurogenesis defects in onset and progression. Cell Mol Life Sci 72(4):773–97. doi:10.1007/s00018-014-1774-1
Grespi F, Melino G (2012) P73 and age-related diseases: is there any link with Parkinson disease? Aging (Albany NY) 4(12):923–31
Garcia-Reitboeck P, Anichtchik O, Dalley JW, Ninkina N, Tofaris GK, Buchman VL, Spillantini MG (2013) Endogenous alpha-synuclein influences the number of dopaminergic neurons in mouse substantia nigra. Exp Neurol 248:541–5. doi:10.1016/j.expneurol.2013.07.015
Nicklas S, Otto A, Wu X, Miller P, Stelzer S, Wen Y, Kuang S, Wrogemann K et al (2012) TRIM32 regulates skeletal muscle stem cell differentiation and is necessary for normal adult muscle regeneration. PLoS One 7(1):e30445. doi:10.1371/journal.pone.0030445
Yang A, Walker N, Bronson R, Kaghad M, Oosterwegel M, Bonnin J, Vagner C, Bonnet H et al (2000) p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature 404(6773):99–103. doi:10.1038/35003607
Bahnassawy L, Perumal TM, Gonzalez-Cano L, Hillje AL, Taher L, Makalowski W, Suzuki Y, Fuellen G et al (2015) TRIM32 modulates pluripotency entry and exit by directly regulating Oct4 stability. Sci Rep 5:13456. doi:10.1038/srep13456
Vicario-Abejon C, Yusta-Boyo MJ, Fernandez-Moreno C, de Pablo F (2003) Locally born olfactory bulb stem cells proliferate in response to insulin-related factors and require endogenous insulin-like growth factor-I for differentiation into neurons and glia. J Neurosci 23(3):895–906
Gonzalez-Cano L, Herreros-Villanueva M, Fernandez-Alonso R, Ayuso-Sacido A, Meyer G, Garcia-Verdugo JM, Silva A, Marques MM et al (2010) p73 deficiency results in impaired self renewal and premature neuronal differentiation of mouse neural progenitors independently of p53. Cell Death Dis 1:e109. doi:10.1038/cddis.2010.87
Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP (2003) Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 31(4):e15
Acknowledgments
The authors would like to thank Dr. Leonidas Stefanis and Dr. Daniel Haber for plasmids as well as Thea van Wuellen and Inga Werthschulte for excellent technical assistance.
Author Contributions
MASP did the conception and design, collection and assembly of data, data analysis and interpretation, manuscript writing and final approval of the manuscript. NC and JG did the model design and interpretation. SFA, LGC and MCM did the collection and assembly of data, data analysis and interpretation. SN did the generation of mNSCs lines. JCS did the conception and design, financial support, data analysis and interpretation, manuscript writing and final approval of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
The J. C. S.’s lab is supported by the Boehringer Ingelheim Foundation and the fund “Innovative Medical Research” of the University of Münster Medical School, Schram-Stiftung (T287/21795/2011) by the Fonds National de la Recherche (FNR) Luxembourg (CORE, C13/BM/5791363), a University Luxembourg Internal Research Project (MidNSCs) and the EU Joint Programme - Neurodegenerative Disease Research (JPND) project (supported by the FNR). L.G.C. was supported by a fellowship from the FNR (AFR, Aides à la Formation-Recherche). M.C.M ’s lab is supported by Grant SAF2012-36143 from Spanish Ministerio de Ciencia e and LE310U14 from the Junta de Castilla y Leon. S.F.A holds a predoctoral contract (PIRTU) from Junta de Castilla y Leon.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Online Resource 1
TRIM32 is localised in the nucleus of neurons of the mouse olfactory bulb and in differentiated mouse neuronal cells. a Immunostainings of mouse brain sections labelled with the indicated antibodies. Images were taken in the olfactory bulb (OB). Scale bars = 20 μm. b Immunostainings of mouse neural stem cells submitted to 5-day differentiation, labelled with the indicated antibodies. Scale bars = 20 μm (PDF 6935 kb)
Online Resource 2
Investigation of possible links in the regulatory network involving trim32 and snca The plots show the difference between experimental data (coloured stars) and simulations (black dots). For each combination of input/output genes a model has been trained using the experimental data and then used to simulate the gene response. The FIT between simulation and data can be used as a measure of the strength of the corresponding causal link. a, b Analysis of other possible regulatory effects of trim32 on a neuronal differentiation-associated genes (map2, neuroD, rbfox3) and b the regulatory effects of the same genes on snca. c, d Analysis of other possible regulatory effects of trim32 on c housekeeping genes (chmpa2, gpi1, psmb2, rab7) and d the regulatory effects of the same genes on snca. (PDF 445 kb)
Online Resource 3
TRIM32 is inducing or inhibiting the transcriptional activity of snca in a concentration-dependent manner, in HEK293T cells Diagram showing the normalised luciferase activity, in the presence of TRIM32 overexpression; 1.33 μg of TRIM32 were coexpressed together with 1.33 μg of an empty vector and 1.33 μg of the luciferase constructs (basic pGL3 vector or pGL3/snca intron 1) in HEK293T cells (mean ± SD; n = 6 independent experiments; Mann-Whitney U test, *p ≤ 0.05); 1.75 μg of TRIM32 were coexpressed together with 0.75 μg of an empty vector and 1.75 μg of the luciferase constructs (basic pGL3 vector or pGL3/snca intron 1) in HEK293T cells (mean ± SD; n = 6 independent experiments; Mann-Whitney U test, *p ≤ 0.05); 2.0 μg of TRIM32 were coexpressed together with 2.0 μg of the luciferase constructs (basic pGL3 vector or pGL3/snca intron 1) in HEK293T cells (mean ± SD; n = 5 independent experiments; t test, *p ≤ 0.05) (PDF 338 kb)
Online Resource 4
ChIP results indicating interaction of TRIM32 with the snca promoter region. Original agarose gel electrophoresis showing PCR results from ChIP-eluted DNA, with representative primer pairs of snca promoter region. A 100-bp DNA ruler was used on the first lane of both upper and lower images and the samples are indicated. (PDF 537 kb)
Online Resource 5
TRIM32 interaction with the snca promoter region. RT-qPCR results showing the DNA fold enrichment under different immunoprecipitations. RT-qPCR was performed on the eluted DNA derived from chromatin immunoprecipitations either with the IgG (negative control), histone 3 (H3, positive control), or TRIM32 antibodies. Results from three primer pairs are shown, depicting the possible interaction of H3 or TRIM32 relative to IgG, (mean ± SD; n = 3 independent experiments; t test, *p ≤ 0.05, **p ≤ 0.001). Fold enrichment was calculated by using the formula 2-DDCt, where DDCt is (Ct H3 or TRIM32) – (Ct IgG)) (PDF 326 kb)
Online Resource 6
Interaction with the snca promoter region is abolished when TRIM32 is knocked down. a, b RT-qPCR results of eluted DNA derived from three independent ChIP experiments, where a shRNA scrambled (a) or a shRNA TRIM32 (b) constructs were used. Five primer pairs are depicting the fold enrichment of TRIM32 compared to IgG (mean ± SD; n = 3 independent experiments; t test, *p ≤ 0.05). Fold enrichment was calculated by using the formula 2−DDCt, where DDCt is (Ct TRIM32) – (Ct IgG) (PDF 177 kb)
Online Resource 7
mRNA levels of trim32 on wt and p73 ko mNSCs are significantly different on the transition from maintenance towards differentiation conditions. a RT-qPCR measuring the relative trim32 mRNA expression levels in wt and TRIM32 ko mNSCs. Values were normalised to GAPDH levels, (mean ± SD; n = 8 independent experiments with n = 4 different cell lines; Mann-Whitney U test, *p ≤ 0.05, **p ≤ 0.001). b RT-qPCR measuring the relative trim32 mRNA expression levels in wt mNSCs under maintenance conditions or after 5 days induction of neuronal differentiation. Note how trim32 mRNA levels are significantly increased on the transition from maintenance towards differentiation. Values were normalised to GAPDH levels, (mean ± SD; n = 4 independent experiments with n = 4 different cell lines; Mann-Whitney U test, *p ≤ 0.05). c RT-qPCR measuring the relative trim32 mRNA expression levels in wt or p73 ko mNSCs, under maintenance conditions and when cells were submitted for 1, 3 or 5 days of neuronal differentiation. Note how trim32 mRNA levels were increased in the mNSCs of both genotypes, though the rate of increase is significantly lower in the p73 ko cells. Values were normalised to 18S levels, (mean ± SD; n ≥ 3 independent experiments; t test, *p ≤ 0.05, **p ≤ 0.001), maint maintenance. (PDF 160 kb)
Online Resource 8
mRNA levels of snca between wt and TRIM32 ko or p73 ko MEFs reveal dissimilarities. a RT-qPCR measuring the relative trim32 mRNA expression levels in wt and TRIM32 ko MEFs. Values were normalised to GAPDH levels, (mean ± SD; n = 3 independent experiments with n = 3 different cell lines; Mann-Whitney U test, *p ≤ 0.05, **p ≤ 0.001). b RT-qPCR measuring the relative snca mRNA expression levels in wt and TRIM32 ko MEFs. The levels of snca mRNA do not seem to change between the two different genotypes, supporting the hypothesis of a cell type-specific effect of snca. Values were normalised to GAPDH levels, (mean ± SD; n = 3 independent experiments with n = 3 different cell lines; Mann-Whitney U test, *p ≤ 0.05). c, d RT-qPCR measuring the relative trim32 and scna mRNA expression levels in wt and p73 ko MEFs. Absence of p73 influences the mRNA levels of both genes validating for the former already published results [22] and revealing a novel effect on the latter possibly attributed to the downregulation of trim32. Values were normalised to 18S levels, (mean ± SD; n ≥3 independent experiments with n = 4 different clones; t test, *p ≤ 0.05, **p ≤ 0.001). (PDF 154 kb)
Rights and permissions
About this article
Cite this article
Pavlou, M.A.S., Colombo, N., Fuertes-Alvarez, S. et al. Expression of the Parkinson’s Disease-Associated Gene Alpha-Synuclein is Regulated by the Neuronal Cell Fate Determinant TRIM32. Mol Neurobiol 54, 4257–4270 (2017). https://doi.org/10.1007/s12035-016-9989-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-016-9989-9