Abstract
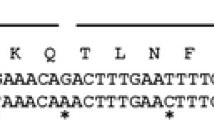
The auxiliary activity family 9 (AA9, formerly GH61) harbors a recently discovered group of oxidative enzymes that boost cellulose degradation. Indeed, these lytic polysaccharide monooxygenases (LPMOs) are able to disrupt the crystalline structure of cellulose, thereby facilitating the work of hydrolytic enzymes involved in biomass degradation. Since these enzymes require an N-terminal histidine residue for activity, their recombinant production as secreted protein is not straightforward. We here report the expression optimization of Trichoderma reesei Cel61A (TrCel61A) in the host Pichia pastoris. The use of the native TrCel61A secretion signal instead of the alpha-mating factor from Saccharomyces cerevisiae was found to be crucial, not only to obtain high protein yields (>400 mg/L during fermentation) but also to enable the correct processing of the N-terminus. Furthermore, the LPMO activity of the enzyme is demonstrated here for the first time, based on its degradation profile of a cellulosic substrate.




Similar content being viewed by others
References
Vu, V. V., Beeson, W. T., Span, E. A., et al. (2014). A family of starch-active polysaccharide monooxygenases. Proc Natl Acad Sci USA, 111, 13822–13827.
Levasseur, A., Drula, E., Lombard, V., et al. (2013). Expansion of the enzymatic repertoire of the CAZy database to integrate auxiliary redox enzymes. Biotechnol Biofuels, 6, 41.
Lo Leggio, L., Simmons, T. J., Poulsen, J.-C. N., et al. (2015). Structure and boosting activity of a starch-degrading lytic polysaccharide monooxygenase. Nat Commun, 6, 5961.
Horn, S. J., Vaaje-Kolstad, G., Westereng, B., et al. (2012). Novel enzymes for the degradation of cellulose. Biotechnol Biofuels, 5, 45.
Dimarogona, M., Topakas, E., Olsson, L., et al. (2012). Lignin boosts the cellulase performance of a GH-61 enzyme from Sporotrichum thermophile. Bioresour Technol, 110, 480–487.
Beeson, W. T., Phillips, C. M., Cate, J. H. D., et al. (2012). Oxidative cleavage of cellulose by fungal copper-dependent polysaccharide monooxygenases. J Am Chem Soc, 134, 890–892.
Li, X., Beeson, W. T., Phillips, C. M., et al. (2012). Structural basis for substrate targeting and catalysis by fungal polysaccharide monooxygenases. Structure, 20, 1051–1061.
Ekwe, E., Morgenstern, I., Tsang, A., et al. (2013). Non-hydrolytic cellulose active proteins: research progress and potential application in biorefineries. Ind Biotechnol, 9, 123–131.
Dimarogona, M., Topakas, E., & Christakopoulos, P. (2013). Recalcitrant polysaccharide degradation by novel oxidative biocatalysts. Appl Microbiol Biotechnol, 97, 8455–8465.
Karkehabadi, S., Hansson, H., Kim, S., et al. (2008). The first structure of a glycoside hydrolase family 61 member, Cel61B from Hypocrea jecorina, at 1.6 A resolution. J Mol Biol, 383, 144–154.
Westereng, B., Ishida, T., Vaaje-Kolstad, G., et al. (2011). The putative endoglucanase PcGH61D from Phanerochaete chrysosporium is a metal-dependent oxidative enzyme that cleaves cellulose. PLoS ONE, 6, e27807.
Kittl, R., Kracher, D., Burgstaller, D., et al. (2012). Production of four Neurospora crassa lytic polysaccharide monooxygenases in Pichia pastoris monitored by a fluorimetric assay. Biotechnol Biofuels, 5, 79.
Bey, M., Zhou, S., Poidevin, L., et al. (2013). Cello-oligosaccharide oxidation reveals differences between two lytic polysaccharide monooxygenases (family GH61) from Podospora anserina. Appl Environ Microbiol, 79, 488–496.
Daly, R., & Hearn, M. T. W. (2005). Expression of heterologous proteins in Pichia pastoris: a useful experimental tool in protein engineering and production. J Mol Recognit, 18, 119–138.
Quinlan, R. J., Sweeney, M. D., Lo Leggio, L., et al. (2011). Insights into the oxidative degradation of cellulose by a copper metalloenzyme that exploits biomass components. Proc Natl Acad Sci USA, 108, 15079–15084.
Hegde, R. S., & Bernstein, H. D. (2006). The surprising complexity of signal sequences. Trends Biochem Sci, 31, 563–571.
Merino, S. T., Cherry, J., & Ave, D. (2007). Progress and challenges in enzyme development for biomass utilization. Adv Biochem Eng Biotechnol, 108, 95–120.
Wilson, D. B. (2009). Cellulases and biofuels. Curr Opin Biotechnol, 20, 295–299.
Kubicek, C. P., Mikus, M., Schuster, A., et al. (2009). Metabolic engineering strategies for the improvement of cellulase production by Hypocrea jecorina. Biotechnol Biofuels, 2, 19.
Saloheimo, M., Nakari-Setälä, T., Tenkanen, M., et al. (1997). cDNA cloning of a Trichoderma reesei cellulase and demonstration of endoglucanase activity by expression in yeast. Eur J Biochem, 249, 584–591.
Karlsson, J., Saloheimo, M., Siika-Aho, M., et al. (2001). Homologous expression and characterization of Cel61A (EG IV) of Trichoderma reesei. Eur J Biochem, 268, 6498–6507.
Näätsaari, L., Mistlberger, B., Ruth, C., et al. (2012). Deletion of the pichia pastoris ku70 homologue facilitates platform strain generation for gene expression and synthetic biology. PLoS ONE, 7, e39720.
Sanchis, J., Fernández, L., Carballeira, J. D., et al. (2008). Improved PCR method for the creation of saturation mutagenesis libraries in directed evolution: application to difficult-to-amplify templates. Appl Microbiol Biotechnol, 81, 387–397.
Gibson, D. G., Young, L., Chuang, R., et al. (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases., 6, 12–16.
Lin-Cereghino, J., Wong, W. W., Xiong, S., et al. (2005). Condensed protocol for competent cell preparation and transformation of the methylotrophic yeast Pichia pastoris. Biotechniques, 38, 44–48.
Weis, R., Luiten, R., Skranc, W., et al. (2004). Reliable high-throughput screening with Pichia pastoris by limiting yeast cell death phenomena. FEMS Yeast Res, 5, 179–189.
De Winter, K., Šimčíková, D., Schalck, B., et al. (2013). Chemoenzymatic synthesis of α-l-rhamnosides using recombinant α-l-rhamnosidase from Aspergillus terreus. Bioresour Technol, 147, 640–644.
Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685.
Sun, L. W., Zhao, Y., Niu, L. P., et al. (2011). A rapid method for determining the concentration of recombinant protein secreted from Pichia pastoris. J Phys: Conf Ser, 276, 012144.
Wood, T. M. (1988). Preparation of crystalline, amorphous, and dyed cellulase substrates. Methods Enzymol, 160, 19–25.
Forsberg, Z., Vaaje-Kolstad, G., Westereng, B., et al. (2011). Cleavage of cellulose by a CBM33 protein. Protein Sci, 20, 1479–1483.
Cregg, J. M., Cereghino, J. L., Shi, J., et al. (2000). Recombinant protein expression in Pichia pastoris. Mol Biotechnol, 16, 23–52.
Hohenblum, H., Gasser, B., Maurer, M., et al. (2004). Effects of gene dosage, promoters, and substrates on unfolded protein stress of recombinant Pichia pastoris. Biotechnol Bioeng, 85, 367–375.
Mellitzer, A., Weis, R., Glieder, A., et al. (2012). Expression of lignocellulolytic enzymes in Pichia pastoris. Microb Cell Fact, 11, 61.
Sygmund, C., Kracher, D., Scheiblbrandner, S., et al. (2012). Characterization of the two Neurospora crassa cellobiose dehydrogenases and their connection to oxidative cellulose degradation. Appl Environ Microbiol, 78, 6161–6171.
Cereghino, J. L., & Cregg, J. M. (2000). Heterologous protein expression in the methylotrophic yeast Pichia pastoris. FEMS Microbiol Rev, 24, 45–66.
Govindappa, N., Hanumanthappa, M., Venkatarangaiah, K., et al. (2014). A new signal sequence for recombinant protein secretion in Pichia pastoris. J Microbiol Biotechnol, 24, 337–345.
Isaksen, T., Westereng, B., Aachmann, F. L., et al. (2014). A C4-oxidizing lytic polysaccharide monooxygenase cleaving both cellulose and cello-oligosaccharides. J Biol Chem, 289, 2632–2642.
Vu, V. V., Beeson, W. T., Phillips, C. M., et al. (2014). Determinants of regioselective hydroxylation in the fungal polysaccharide monooxygenases. J Am Chem Soc, 136, 562–565.
Forsberg, Z., Mackenzie, A. K., Sorlie, M., et al. (2014). Structural and functional characterization of a conserved pair of bacterial cellulose-oxidizing lytic polysaccharide monooxygenases. Proc Natl Acad Sci, 111, 8446–8451.
Ahmad, M., Hirz, M., Pichler, H., et al. (2014). Protein expression in Pichia pastoris: recent achievements and perspectives for heterologous protein production. Appl Microbiol Biotechnol, 98, 5301–5317.
Acknowledgments
The authors wish to thank the Agency for Innovation by Science and Technology (IWT) Flanders for financial support (Ph.D.-grant to M.T.). The study was carried out in the frame of the MRP Project “Ghent Bio-economy” granted by Ghent University.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Tanghe, M., Danneels, B., Camattari, A. et al. Recombinant Expression of Trichoderma reesei Cel61A in Pichia pastoris: Optimizing Yield and N-terminal Processing. Mol Biotechnol 57, 1010–1017 (2015). https://doi.org/10.1007/s12033-015-9887-9
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-015-9887-9