Abstract
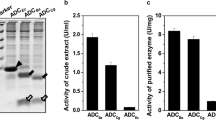
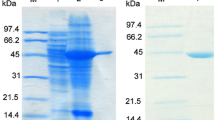
N-Succinyl-amino acid racemase (NSAAR), long referred to as N-acyl- or N-acetyl-amino acid racemase, is an enolase superfamily member whose biotechnological potential was discovered decades ago, due to its use in the industrial dynamic kinetic resolution methodology first known as “Acylase Process”. In previous works, an extended and enhanced substrate spectrum of the NSAAR from Geobacillus kaustophilus CECT4264 toward different N-substituted amino acids was reported. In this work, we describe the cloning, purification, and characterization of the NSAAR from Geobacillus stearothermophilus CECT49 (GstNSAAR). The enzyme has been extensively characterized, showing a higher preference toward N-formyl-amino acids than to N-acetyl-amino acids, thus confirming that the use of the former substrates is more appropriate for a biotechnological application of the enzyme. The enzyme showed an apparent thermal denaturation midpoint of 77.0 ± 0.1 °C and an apparent molecular mass of 184 ± 5 kDa, suggesting a tetrameric species. Optimal parameters for the enzyme activity were pH 8.0 and 55–65 °C, with Co2+ as the most effective cofactor. Mutagenesis and binding experiments confirmed K166, D191, E216, D241, and K265 as key residues in the activity of GstNSAAR, but not indispensable for substrate binding.





Similar content being viewed by others
Notes
We will use the three names indistinctly during this work, to try to maintain the original nomenclature used in the corresponding papers.
References
May, O., Verseck, S., Bommarius, A., & Drauz, K. (2002). Development of dynamic kinetic resolution processes for biocatalytic production of natural and nonnatural l-amino acids. Organic Process Research & Development, 6, 452–457.
Tokuyama, S., Hatano, K., & Takahashi, T. (1994). Discovery of a novel enzyme, N-acylamino acid racemase in an actinomycete: Screening, isolation and identification. Bioscience, Biotechnology, and Biochemistry, 58, 24–27.
Tokuyama, S., Miya, H., Hatano, K., & Takahashi, T. (1994). Purification and properties of a novel enzyme, N-acylamino acid racemase, from Streptomyces atratus Y-53. Applied Microbiology and Biotechnology, 40, 835–840.
Tokuyama, S., & Hatano, K. (1995). Purification and properties of thermostable N-acylamino acid racemase from Amycolatopsis sp. TS-1-60. Applied Microbiology and Biotechnology, 42, 853–859.
Tokuyama, S., & Hatano, K. (1995). Cloning, DNA sequencing and heterologous expression of the gene for thermostable N-acylamino acid racemase from Amycolatopsis sp. TS-1-60 in Escherichia coli. Applied Microbiology and Biotechnology, 42, 884–889.
Verseck, S., Bommarius, A., & Kula, M. R. (2001). Screening, overexpression and characterization of an N-acylamino acid racemase from Amycolatopsis orientalis subsp. lurida. Applied Microbiology and Biotechnology, 55, 354–361.
Su, S.-C., & Lee, C.-Y. (2002). Cloning of the N-acylamino acid racemase gene from Amycolatopsis azurea and biochemical characterization of the gene product. Enyzme and Microbial Technology, 30, 647–655.
Wang, W. C., Chiu, W. C., Hsu, S. K., Wu, C. L., Chen, C. Y., Liu, J. S., & Hsu, W. H. (2004). Structural basis for catalytic racemization and substrate specificity of an N-acylamino acid racemase homologue from Deinococcus radiodurans. Journal of Molecular Biology, 342, 155–169.
Pozo-Dengra, J., Martinez-Gomez, A. I., Martinez-Rodriguez, S., Clemente-Jimenez, J. M., Rodriguez-Vico, F., & Las Heras-Vazquez, F. J. (2009). Racemization study on different N-acetylamino acids by a recombinant N-succinylamino acid racemase from Geobacillus kaustophilus CECT4264. Process Biochemistry, 44, 835–841.
Palmer, D. R., Garrett, J. B., Sharma, V., Meganathan, R., Babbitt, P. C., & Gerlt, J. A. (1999). Unexpected divergence of enzyme function and sequence: “N-acylamino acid racemase” is o-succinylbenzoate synthase. Biochemistry, 38, 4252–4258.
Thompson, T. B., Garrett, J. B., Taylor, E. A., Meganathan, R., Gerlt, J. A., & Rayment, I. (2000). Evolution of enzymatic activity in the enolase superfamily: Structure of o-succinylbenzoate synthase from Escherichia coli in complex with Mg2+ and o-succinylbenzoate. Biochemistry, 39, 10662–10676.
Schmidt, D. M., Hubbard, B. K., & Gerlt, J. A. (2001). Evolution of enzymatic activities in the enolase superfamily: Functional assignment of unknown proteins in Bacillus subtilis and Escherichia coli as l-Ala-D/l-Glu epimerases. Biochemistry, 4051, 15707–15715.
Taylor Ringia, E. A., Garrett, J. B., Thoden, J. B., Holden, H. M., Rayment, I., & Gerlt, J. A. (2004). Evolution of enzymatic activity in the enolase superfamily: Functional studies of the promiscuous o-succinylbenzoate synthase from Amycolatopsis. Biochemistry, 43, 224–229.
Thoden, J. B., Taylor Ringia, E. A., Garrett, J. B., Gerlt, J. A., Holden, H. M., & Rayment, I. (2004). Evolution of enzymatic activity in the enolase superfamily: Structural studies of the promiscuous o-succinylbenzoate synthase from Amycolatopsis. Biochemistry, 43, 5716–5727.
Glasner, M. E., Fayazmanesh, N., Chiang, R. A., Sakai, A., Jacobson, M. P., Gerlt, J. A., & Babbitt, P. C. (2006). Evolution of structure and function in the o-succinylbenzoate synthase/N-acylamino acid racemase family of the enolase superfamily. Journal of Molecular Biology, 360, 228–250.
Sakai, A., Xiang, D. F., Xu, C., Song, L., Yew, W. S., Raushel, F. M., & Gerlt, J. A. (2006). Evolution of enzymatic activities in the enolase superfamily: N-succinylamino acid racemase and a new pathway for the irreversible conversion of d- to l-amino acids. Biochemistry, 45, 4455–4462.
Song, L., Kalyanaraman, C., Fedorov, A. A., Fedorov, E. V., Glasner, M. E., Brown, S., et al. (2007). Prediction and assignment of function for a divergent N-succinyl amino acid racemase. Nature Chemical Biology, 3, 486–491.
Bommarius, A., Drauz, K., Kula, M.-R., & Verseck, S. (2001). N-Acetylamino acid racemase. EP 1074628 A1.
Bommarius, A., Drauz, K., Verseck, S., & Kula, M.-R. (2004). Acetyl amino acid racemase from Amycolatopsis orientalis for racemizing carbamoyl amino acids. US Patent 6767725 B2.
Bommarius, A., Drauz, K., & Verseck, S. (2008). Racemization and deprotection of special N-protected amino acids in the acylase/racemase system for the total conversion of special N-protected racemic amino acids into optically pure amino acids. US Patent 7378269 B2.
Verseck, S., Kula, M.-R, Bommarius, A., & Drauz, K. (2002). For producing enantiomer-enriched amino acids, and derivatives. US Patent 6372459 B1.
Takahashi, T., & Hatano, K. (1989). Acylamino acid racemase, Production and use thereof. EP 0304021 A2.
Takahashi, T., & Hatano, K. (1991). Acylamino acid racemase, production and use thereof. US Patent 4981799 A.
Tokuyama, M., Hatano, K., Nakahama, K., & Takahashi, T. (1992). DNA encoding acylamino acid racemase and its use. EP 0474965 A2.
Srivibool, R., Kurakami, K., Sukchotiratana, M., & Tokuyama, S. (2004). Coastal soil actinomycetes: Thermotolerant strains producing N-acylamino acid racemase. Science Asia, 30, 123–126.
Tokuyama, S., & Hatano, K. (1996). Overexpression of the gene for N-acylamino acid racemase from Amycolatopsis sp. TS-1-60 in Escherichia coli and continuous production of optically active methionine by a bioreactor. Applied Microbiology and Biotechnology, 44, 774–777.
Chiu, W. C., You, J. Y., Liu, J. S., Hsu, S. K., Hsu, W. H., Shih, C. H., et al. (2006). Structure–stability–activity relationship in covalently cross-linked N-carbamoyl-d-amino acid amidohydrolase and N-acylamino acid racemase. Journal of Molecular Biology, 359, 741–753.
Hsu, S. K., Lo, H. H., Kao, C. H., Lee, D. S., & Hsu, W. H. (2006). Enantioselective synthesis of l-homophenylalanine by whole cells of recombinant Escherichia coli expressing l-aminoacylase and N-acylamino acid racemase genes from Deinococcus radiodurans BCRC12827. Biotechnology Progress, 22, 1578–1584.
Hsu, S., Lo, H., Lin, W., Chen, I., Kao, C., & Hsu, W. (2007). Stereoselective synthesis of l-homophenylalanine using the carbamoylase method with in situ racemization via N-acylamino acid racemase. Process Biochemistry, 42, 856–862.
Yen, M.-C., Hsu, W.-H., & Lin, S.-C. (2010). Synthesis of l-homophenylalanine with immobilized enzymes. Process Biochemistry, 45, 667–674.
Pozo-Dengra, J., Martínez-Rodríguez, S., Contreras, L. M., Prieto, J., Andújar-Sánchez, M., Clemente-Jiménez, J. M., et al. (2009). Structure and conformational stability of a tetrameric thermostable N-succinylamino acid racemase. Biopolymers, 91, 757–772.
Soriano-Maldonado, P., Rodríguez-Alonso, M. J., Hernández-Cervantes, C., Rodríguez-García, I., Clemente-Jiménez, J. M., Rodríguez-Vico, F., et al. (2014). Amidohydrolase process: Expanding the use of l-N-carbamoylase/N-succinyl-amino acid racemase tandem for the production of different optically pure l-amino acids. Process Biochemistry, 49, 1281–1287.
Soriano-Maldonado, P., Las Heras-Vazquez, F.J., Clemente-Jimenez, J.M., Rodriguez-Vico, F., & Martínez-Rodríguez, S. (2014). Enzymatic dynamic kinetic resolution of racemic N-formyl- and N-carbamoyl-amino acids using immobilized l-N-carbamoylase and N-succinyl-amino acid racemase. Applied Microbiology and Biotechnology. doi:10.1007/s00253-014-5880-7.
Hayashida, M., Kim, S. H., Takeda, K., Hisano, T., & Miki, K. (2008). Crystal structure of N-acylamino acid racemase from Thermus thermophilus HB8. Proteins, 71, 519–523.
Baxter, S., Royer, S., Grogan, G., Brown, F., Holt-Tiffin, K. E., Taylor, I. N., et al. (2012). An improved racemase/acylase biotransformation for the preparation of enantiomerically pure amino acids. Journal of the American Chemical Society, 134, 19310–19313.
Martínez-Rodríguez, S., Martínez-Gómez, A. I., Rodríguez-Vico, F., Clemente-Jiménez, J. M., & Las Heras-Vázquez, F. J. (2010). N-Carbamoyl-d- and l-amino acid amidohydrolases: Characteristics and applications in biotechnological processes. Applied Microbiology and Biotechnology, 85, 441–458.
Stumpp, T., Wilms, B., & Altenbuchner, J. (2000). Ein neues, l-rhamnoseinduzierbares expressionssystem für Escherichia coli. Biospektrum, 6, 33–36.
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics, 23, 2947–2948.
Gill, S. C., & von Hippel, P. H. (1989). Calculation of protein extinction coefficients from amino acid sequence data. Analytical Biochemistry, 182, 319–326.
Zhu, W. W., Wang, C., Jipp, J., Ferguson, L., Lucas, S. N., Hicks, M. A., & Glasner, M. E. (2012). Residues required for activity in Escherichia coli o-succinylbenzoate synthase (OSBS) are not conserved in all OSBS enzymes. Biochemistry, 51, 6171–6181.
Babbitt, P. C., Hasson, M. S., Wedekind, J. E., Palmer, D. R., Barrett, W. C., Reed, G. H., et al. (1996). The enolase superfamily: A general strategy for enzyme-catalyzed abstraction of the alpha-protons of carboxylic acids. Biochemistry, 35, 16489–16501.
Galisteo, M. L., Mateo, P. L., & Sánchez-Ruiz, J. M. (1991). Kinetic study on the irreversible thermal denaturation of yeast phosphoglycerate kinase. Biochemistry, 30, 2061–2066.
Martínez-Rodríguez, S., Encinar, J. A., Hurtado-Gómez, E., Prieto, J., Clemente-Jiménez, J. M., Las Heras-Vázquez, F. J., et al. (2009). Metal-triggered changes in the stability and secondary structure of a tetrameric dihydropyrimidinase: A biophysical characterization. Biophysical Chemistry, 139, 42–52.
Eisenthal, R., Peterson, M. E., Daniel, R. M., & Danson, M. J. (2006). The thermal behaviour of enzyme activity: Implications for biotechnology. Trends in Biotechnology, 24, 289–292.
Daniel, R. M., Peterson, M. E., Danson, M. J., Price, N. C., Kelly, S. M., Monk, C. R., et al. (2009). The molecular basis of the effect of temperature on enzyme activity. Biochemical Journal, 425, 3533–3560.
Odokonyero, D., Ragumani, S., Lopez, M. S., Bonanno, J. B., Ozerova, N. D., Woodard, D. R., et al. (2013). Divergent evolution of ligand binding in the o-succinylbenzoate synthase family. Biochemistry, 52, 7512–7521.
McMillan, A. W., Lopez, M. S., Zhu, M., Morse, B. C., Yeo, I. C., Amos, J., et al. (2014). Role of an active site loop in the promiscuous activities of Amycolatopsis sp. T-1-60 NSAR/OSBS. Biochemistry, 53, 4434–4444.
Acknowledgments
We thank Andy Taylor for critical discussion of the manuscript and Pedro Madrid-Romero for technical assistance. This work was supported by the Spanish Ministry of Education and Science, the European Social Fund (ESF), and the European Regional Development Fund (ERDF), through the project BIO2011-27842, by the Andalusian Regional Council of Innovation, Science and Technology, through the project TEP-4691, and by the European Cooperation in Science and Technology (COST) Action CM1303. P.S.-M. was supported by the University of Almería. S.M.-R. was supported by the Spanish Ministry of Science and Innovation.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Soriano-Maldonado, P., Andújar-Sánchez, M., Clemente-Jiménez, J.M. et al. Biochemical and Mutational Characterization of N-Succinyl-Amino Acid Racemase from Geobacillus stearothermophilus CECT49. Mol Biotechnol 57, 454–465 (2015). https://doi.org/10.1007/s12033-015-9839-4
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-015-9839-4