Abstract
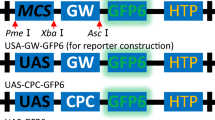
The genome information is offering opportunities to manipulate genes, polygenic characters and multiple traits in plants. Although a number of approaches have been developed to manipulate traits in plants, technical hurdles make the process difficult. Gene cloning vectors that facilitate the fusion, overexpression or down regulation of genes in plant cells are being used with various degree of success. In this study, we modified gateway MultiSite cloning vectors and developed a hybrid cloning strategy which combines advantages of both traditional cloning and gateway recombination cloning. We developed Gateway entry (pGATE) vectors containing attL sites flanking multiple cloning sites and plant expression vector (pKM12GW) with specific recombination sites carrying different plant and bacterial selection markers. We constructed a plant expression vector carrying a reporter gene (GUS), two Bt cry genes in a predetermined pattern by a single round of LR recombination reaction after restriction endonuclease-mediated cloning of target genes into pGATE vectors. All the three transgenes were co-expressed in Arabidopsis as evidenced by gene expression, histochemical assay and insect bioassay. The pGATE vectors can be used as simple cloning vectors as there are rare restriction endonuclease sites inserted in the vector. The modified multisite vector system developed is ideal for stacking genes and pathway engineering in plants.







Similar content being viewed by others
References
Jacobsen, E., & Nataraja, K. N. (2008). Cisgenics—facilitating the second green revolution in India by improved traditional plant breeding. Current Science, 94, 1365–1366.
Capell, T., & Christou, P. (2004). Progress in plant metabolic engineering. Current Opinion in Biotechnology, 15, 148–154.
Dafny-Yelin, M., & Tzfira, T. (2007). Delivery of multiple transgenes to plant cells. Plant Physiology, 145, 1118–1128.
Halpin, C. (2005). Gene stacking in transgenic plants—the challenge for 21st century plant biotechnology. Plant Biotechnology Journal, 3, 141–155.
Naqvi, S., Farre′, G., Sanahuja, G., Capell, T., Zhu, C., & Christou, P. (2010). When more is better: Multigene engineering in plants. Trends in Plant Science, 15, 48–56.
Zhao, J. Z., Cao, J., Li, Y., Collins, H. L., Roush, R. T., Earle, E. D., et al. (2003). Transgenic plants expressing two Bacillus thuringiensis toxins delays insect resistance evolution. Nature Biotechnology, 21, 1493–1497.
Wu, G., Truska, M., Datla, N., Vrinten, P., Bauer, J., Zank, T., et al. (2005). Step-wise engineering to produce high yields of very long-chain polyunsaturated fatty acids in plants. Nature Biotechnology, 23, 1013–1017.
Seitz, C., Vitten, M., Steinbach, P., Hartl, S., Hirsche, J., Rathje, W., et al. (2007). Redirection of anthocyanin synthesis in Osteospermum hybrida by a two-enzyme manipulation strategy. Phytochemistry, 68, 824–833.
Singla-Pareek, S. L., Reddy, M. K., & Sopory, S. K. (2003). Genetic engineering of the glyoxalase pathway in tobacco leads to enhanced salinity tolerance. Proceedings of the National Academy of Sciences of the United States of America, 100, 14672–14677.
Lin, L., Liu, Y. G., Xu, X., & Li, B. (2003). Efficient linking and transfer of multiple genes by a multigene assembly and transformation vector system. Proceedings of the National Academy of Sciences of the United States of America, 100, 5962–5967.
Lucker, J., Schwab, W., van Hautum, B., Blaas, J., van der Plas, L. H., Bouwmeester, H. J., et al. (2004). Increased and altered fragrance of tobacco plants after metabolic engineering using three monoterpene synthases from lemon. Plant Physiology, 134, 510–519.
Hadi, M. Z., McMullen, M. D., & Finer, J. J. (1996). Transformation of 12 different plasmids into soybean via particle bombardment. Plant Cell Reports, 15, 500–505.
Chen, L., Marmey, P., Taylor, N. J., Brizard, J. P., Espinoza, C., D’Cruz, P., et al. (1998). Expression and inheritance of multiple transgenes in rice plants. Nature Biotechnology, 16, 1060–1064.
Ye, X., Al Babili, S., Kloti, A., Zhang, J., Lucca, P., Beyer, P., et al. (2000). Engineering the provitamin A (β-carotene) biosynthetic pathway into (carotenoid-free) rice endosperm. Science, 287, 303–305.
Nawrath, C., Poirier, Y., & Somerville, C. (1994). Targeting of the polyhydroxybutyrate biosynthetic-pathway to the plastids of Arabidopsis thaliana results in high-levels of polymer accumulation. Proceedings of the National Academy of Sciences of the United States of America, 91, 12760–12764.
Halpin, C., Barakate, A., Askari, B. M., Abbott, J. C., & Ryan, M. D. (2001). Enabling technologies for manipulating multiple genes on complex pathways. Plant Molecular Biology, 47, 295–310.
Daniell, H., & Dhingra, A. (2002). Multigene engineering: Dawn of an exciting new era in biotechnology. Current Opinion in Biotechnology, 13, 136–141.
Ma, C. L., & Mitra, A. (2002). Expressing multiple genes in a single open reading frame with the 2A region of foot-and-mouth disease virus as a linker. Molecular Breeding, 9, 191–199.
Karimi, M., Bleys, A., Vanderhaeghen, R., & Hilson, P. (2007). Building blocks for plant gene assembly. Plant Physiology, 145, 1183–1191.
Walhout, A., Temple, G., Brasch, M., Hartley, J., Lorson, M., van den Heuvel, S., et al. (2000). GATEWAY recombinational cloning: application to the cloning of large numbers of open reading frames or ORFeomes. Methods in Enzymology, 328, 575–592.
Chen, Q. J., Zhou, H. M., Chen, J., & Wang, X. C. (2006). A Gateway-based platform for multigene plant transformation. Plant Molecular Biology, 62, 927–936.
Mahmood. T., Zar. T., & Saqlan Naqvi. S. M. (2008). Multiple pulses improve electroporation efficiency in Agrobacterium tumefaciens. Electronic J Biotechnology. ISSN: 0717-3458 11:1–4.
Clough, S. J., & Bent, A. F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal, 16, 735–743.
Datta, K., Schmidt, A., & Marcus, A. (1989). Characterization of two soybean repetitive proline-rich proteins and a cognate cdna from geminated axes. Plant Cell, 1, 945–952.
Jefferson, R. A., Kavanagh, T. A., & Bevan, M. W. (1987). GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO Journal, 6, 3901–3907.
Sundaram, K. M. S., & Sundaram, A. (1992). An insect bioassay method to determine persistence of Bacillus thuringiensis var. Kurstaki (B.t.k.) protein in oak foliage, following application of a commercial formulation under field and laboratory conditions. Journal of Environmental Science and Health. Part B, 27, 73–112.
Halpin, C., & Boerjan, W. (2003). Stacking transgenes in forest trees. Trends in Plant Science, 8, 363–365.
Curtis, M. D., & Grossniklaus, U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiology, 133, 462–469.
Earley, K. W., Haag, J. R., Pontes, O., Opper, K., Juehne, T., Song, K., et al. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. The Plant Journal, 45, 616–629.
Karimi, M., Inze, D., & Depicker, A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends in Plant Science, 7, 193–195.
Walter, M., Chaban, C., Schutze, K., Batistic, O., Weckermann, K., Nake, C., et al. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. The Plant Journal, 40, 428–438.
Rozwoadowski, K., Yang, W., & Kagale, S. (2008). Homologous recombination-mediated cloning and manipulation of genomic DNA regions using Gateway and recombineering systems. BMC Biotechnology, 8, 88.
Chakrabarthi, S. K., Mandaokar, A. D., Ananda kumar, P., & Sharma, R. P. (1998). Synergistic effect of cry1Ac and cry1F δ-endotoxins of Bacillus thuringiensis on cotton bollworm, Helicoverpa armigera. Current Science, 75, 663–664.
Acknowledgments
This study is supported by the research grants from the Indian Council of Agricultural Research, Department of Biotechnology and DST-FIST Government of India. Authors would like to thank to Dr. Rakesh Tuli, National Agri-Food Biotechnology Institute, Mohali, India and Dr. P. Anandakumar, National Research Centre for Plant Biotechnology, IARI, New Delhi 110012, India, for providing cry1Ec and cry1Aabc genes used in this study. Dr. A.R.V Kumar, Department of Entomology, University of Agricultural Sciences, GKVK, Bangalore, India provided DBM insect larvae for insect bioassay.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Vemanna, R.S., Chandrashekar, B.K., Hanumantha Rao, H.M. et al. A Modified MultiSite Gateway Cloning Strategy for Consolidation of Genes in Plants. Mol Biotechnol 53, 129–138 (2013). https://doi.org/10.1007/s12033-012-9499-6
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-012-9499-6