Abstract
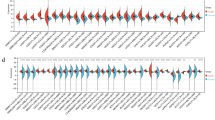
DNA methylation analysis, an epigenetic specification, has been explored for partial determination of cancer cell phenotypes. The development of metastasis in cancerogenesis has led its feasible association with the epigenetic modulations. We generated highly aggressive non-small cell lung cancer cell lines (HTB56 and A549) by using in vivo selection approach. These were, then, subjected to DNA methylation analysis (genome-wide). We also explored the therapeutic effects of azacytidine, an epigenetic agent, on DNA methylation patterns as well as the in vivo phenotypes. During the development of highly aggressive cell lines, we observed widespread modulations in DNA methylation. Reduced representation bisulfite sequencing was used and compared with the less aggressive parental cell lines to identify the differential methylation, which was achieved up to 2.7 % of CpG-rich region. Azacytidine inhibited DNA methyltransferase and reversed the prometastatic phenotype. We found its high association with the preferential loss of DNA methylation from hypermethylated sites. After persisted exposure of azacytidine, we observed that DNA methylation affected the polycomb-binding sites. We found close association of DNA methylome modifications with metastatic capability of non-small cell lung cancer. We also concluded that epigenetic modulation could be used as a potential therapeutic approach to prevent metastasis formation as prometastatic phenotype was reversed due to inhibition of DNA methyltransferase.




Similar content being viewed by others
References
Siegel R, Naishadham D, Jemal A. Cancer statistics. CA Cancer J Clin. 2013;63:11–30.
Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4:143–53.
Konishi K, Watanabe Y, Shen L, et al. DNA methylation profiles of primary colorectal carcinoma and matched liver metastasis. PLoS One. 2011;6:e27889.
Baylin SB, Jones PA. A decade of exploring the cancer epigenome-biological and translational implications. Nat Rev Cancer. 2011;11:726–34.
Geutjes EJ, Bajpe PK, Bernards R. Targeting the epigenome for treatment of cancer. Oncogene. 2012;31:3827–44.
Bjornsson HT, Sigurdsson MI, Fallin MD, et al. Intra-individual change over time in DNA methylation with familial clustering. JAMA. 2008;299:2877–83.
Tsai HC, Li H, Van Neste L, et al. Transient low doses of DNA-demethylating agents exert durable antitumor effects on hematological and epithelial tumor cells. Cancer Cell. 2012;21:430–6.
Raynal NJ, Si J, Taby RF, et al. DNA methylation does not stably lock gene expression but instead serves as a molecular mark for gene silencing memory. Cancer Res. 2012;72:1170–81.
Griffiths EA, Gore SD. Epigenetic therapies in MDS and AML. Adv Exp Med Biol. 2013;754:253–83.
Shats I, Gatza ML, Chang JT, et al. Using a stem cell-based signature to guide therapeutic selection in cancer. Cancer Res. 2011;71:1772–80.
Glinsky GV, Berezovska O, Glinskii AB. Microarray analysis identifies a death-from-cancer signature predicting therapy failure in patients with multiple types of cancer. J Clin Invest. 2005;115:1503–21.
Carmona FJ, Villanueva A, Vidal A, et al. Epigenetic disruption of cadherin-11 in human cancer metastasis. J Pathol. 2012;228:230–40.
Bulk E, Hascher A, Liersch R, et al. Adjuvant therapy with small hairpin RNA interference prevents non-small cell lung cancer metastasis development in mice. Cancer Res. 2008;68:1896–904.
Yu J, Bulk E, Ji P, et al. The EPHB6 receptor tyrosine kinase is a metastasis suppressor that is frequently silenced by promoter DNA hypermethylation in non-small cell lung cancer. Clin Cancer Res. 2010;16:2275–83.
Bulk E, Sargin B, Krug U, et al. S100A2 induces metastasis in non-small cell lung cancer. Clin Cancer Res. 2009;15:22–9.
Bisping G, Wenning D, Kropff M, et al. Bortezomib, dexamethasone, and fibroblast growth factor receptor 3-specific tyrosine kinase inhibitor in t(4;14) myeloma. Clin Cancer Res. 2009;15:520–31.
Kulimova E, Oelmann E, Bisping G, et al. Growth inhibition and induction of apoptosis in acute myeloid leukemia cells by new indolinone derivatives targeting fibroblast growth factor, platelet-derived growth factor, and vascular endothelial growth factor receptors. Mol Cancer Ther. 2006;5:3105–12.
Meissner A, Mikkelsen TS, Gu H, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–70.
Schoofs T, Rohde C, Hebestreit K, et al. DNA methylation changes are a late event in acute promyelocytic leukemia and coincide with loss of transcription factor binding. Blood. 2013;121:178–87.
Fujita PA, Rhead B, Zweig AS, et al. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 2011;39:D876–82.
Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10–2.
Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27:1571–2.
Hebestreit K, Dugas M, Klein HU. Detection of significantly differentially methylated regions in targeted bisulfite sequencing data. Bioinformatics. 2013;29:1647–53.
Hong F, Breitling R, McEntee CW, et al. RankProd: a bioconductor package for detecting differentially expressed genes in meta-analysis. Bioinformatics. 2006;22:2825–57.
Yamamoto G, Nannya Y, Kato M, et al. Highly sensitive method for genomewide detection of allelic composition in nonpaired, primary tumor specimens by use of Affymetrix single-nucleotide-polymorphism genotyping microarrays. Am J Hum Genet. 2007;81:114–26.
Thoennissen NH, Krug UO, Lee DH, et al. Prevalence and prognostic impact of allelic imbalances associated with leukemic transformation of Philadelphia chromosome-negative myeloproliferative neoplasms. Blood. 2010;115:2882–90.
Rius M, Stresemann C, Keller D, et al. Human concentrative nucleoside transporter 1-mediated uptake of 5-azacytidine enhances DNA demethylation. Mol Cancer Ther. 2009;8:225–31.
Rosenbloom KR, Dreszer TR, Pheasant M, et al. ENCODE whole-genome data in the UCSC Genome Browser. Nucleic Acids Res. 2010;38:D620–5.
Thurman RE, Rynes E, Humbert R, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82.
Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–92.
Conflict of interest
None.
Author information
Authors and Affiliations
Corresponding author
Additional information
Shou-Ping Dai and Chao Xie have contributed equally to this work.
Rights and permissions
About this article
Cite this article
Dai, SP., Xie, C., Ding, N. et al. Targeted inhibition of genome-wide DNA methylation analysis in epigenetically modulated phenotypes in lung cancer. Med Oncol 32, 176 (2015). https://doi.org/10.1007/s12032-015-0615-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12032-015-0615-x