Abstract
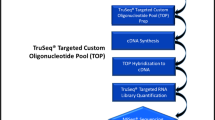
Gene expression investigations are well-established components of ante mortem studies with broad applications ranging from elucidating basic mechanisms responsible for normal physiological processes to discovering therapeutic targets in pathophysiological conditions. However, gene expression studies and their application in the medico-legal field are still in their infancy. Therefore, the present study focuses on RNA using PCR array in the analysis of gene expression associated with tissues taken from actual criminal cases. RNA was extracted from the liver tissues of bodies with PMIs between 6 and 48 h. The results demonstrated that mRNA was stable up to 48 h postmortem. Further, as cell death is an indispensable and necessary part of the biological life cycle, apoptotic gene expression profiles were investigated. The gene expression related to the programmed cell death found in body tissues after death is defined as the apoptotic thanatotranscriptome (thanatos-, Greek for death). On comparison of control and decaying tissues, the results show that with time, pro-apoptotic genes such as caspases are up-regulated and the expression of genes responsible for anti-apoptosis such as BCL2 and BAG3 were down-regulated. Thus, this current work gives a unique perspective of the apoptotic thanatotranscriptome that is affected after death. Up to the present time, gene expression in bodies from criminal cases has not been reported in literature using PCR array techniques. Thus, this thanatotranscriptome study provides insight into postmortem gene activity with potential applications in medico-legal investigations.


Similar content being viewed by others
References
Can I, Javan GT, Pozhitkov AE, Noble PA. Distinctive thanatomicrobiome signatures found in the blood and internal organs of humans. J Microbiol Methods. 2014;106:1–7.
Heinrich M, Matt K, Lutz-Bonengel S, Schmidt U. Successful RNA extraction from various human postmortem tissues. Int J Legal Med. 2007;121:136–42.
Preece P, Cairns NJ. Quantifying mRNA in postmortem human brain: influence of gender, age at death, postmortem interval, brain pH, agonal state and inter-lobe mRNA variance. Mol Brain Res. 2003;118:60–71.
Fitzpatrick R, Casey OM, Morris D, Smith T, Powell R, Sreenan JM. Postmortem stability of RNA isolated from bovine reproductive tissues. Biochim Biophys Acta. 2002;1574:10–4.
Fordyce SL, Kampmann ML, van Doorn NL, Gilbert MT. Long-term RNA persistence in postmortem contexts. Investig Genet. 2013;4:7.
Botling J, Edlund K, Segersten U, Tahmasebpoor S, Engström M, Sundström M, et al. Impact of thawing on RNA integrity and gene expression analysis in fresh frozen tissue. Diagn Mol Pathol. 2009;18:44–52.
Bauer M, Gramlich I, Polzin S, Patzelt D. Quantification of mRNA degradation as possible indicator of postmortem interval—a pilot study. Leg Med. 2003;5:220–7.
Bauer M. RNA in forensic science. Forensic Sci Int Genet. 2007;1:69–74.
Birdsill AC, Walker DG, Lue L, Sue LI, Beach TG. Postmortem interval effect on RNA and gene expression in human brain tissue. Cell Tissue Bank. 2011;12:311–8.
González-Herrera L, Valenzuela A, Marchal JA, Lorente JA, Villanueva E. Studies on RNA integrity and gene expression in human myocardial tissue, pericardial fluid and blood, and its postmortem stability. Forensic Sci Int. 2013;232:218–28.
Finger JM, Mercer JF, Cotton RG, Danks DM. Stability of protein and mRNA in human postmortem liver-analysis by two-dimensional gel electrophoresis. Clin Chim Acta. 1987;170:209–18.
Hansen J, Lesnikova I, Funder AMD, Banner J. DNA and RNA analysis of blood and muscle from bodies with variable postmortem intervals. Forensic Sci Med Pathol. 2014;10:322–8.
Yasojima K, McGeer EG, McGeer PL. High stability of mRNAs postmortem and protocols for their assessment by RT-PCR. Brain Res Protoc. 2001;8:212–8.
Anderson S, Howard B, Hobbs GR, Bishop CP. A method for determining the age of a bloodstain. Forensic Sci Int. 2005;148:37–45.
Bauer M, Polzin S, Patzelt D. Quantification of RNA degradation by semi-quantitative duplex and competitive RT-PCR: a possible indicator of the age of bloodstains? Forensic Sci Int. 2003;138:94–103.
Bauer M, Kraus A, Patzelt D. Detection of epithelial cells in dried blood stains by reverse transcriptase–polymerase chain reaction. J Forensic Sci. 1999;44:1232–6.
Zubakov D, Kokshoorn M, Kloosterman A, Kayser M. New markers for old stains: stable mRNA markers for blood and saliva identification from up to 16-year-old stains. Int J Legal Med. 2009;123:71–4.
Setzer M, Juusola J, Ballantyne J. Recovery and stability of RNA in vaginal swabs and blood, semen, and saliva stains. J Forensic Sci. 2008;53:296–305.
Sakurada K, Ikegaya H, Fukushima H, Akutsu T, Watanabe K, Yoshino M. Evaluation of mRNA-based approach for identification of saliva and semen. Leg Med. 2009;11:125–8.
Kuliwaba JS, Fazzalari NL, Findlay DM. Stability of RNA isolated from human trabecular bone at post-mortem and surgery. Biochim Biophys Acta. 2005;1740:1–11.
van Doorn NL, Wilson AS, Willerslev E, Gilbert MTP. Bone marrow and bone as a source for postmortem RNA. J Forensic Sci. 2011;56:720–5.
King K, Flinter FA, Green PM. Hair roots as the ideal source of mRNA for genetic testing. J Med Genet. 2001;38:e20.
Williams T, Soni S, White J, Can G, Javan GT. Evaluation of DNA degradation using flow cytometry: promising tool for postmortem interval determination. Am J Forensic Med Pathol. 2015;36:104–10.
Sampaio-Silva F, Magalhães T, Carvalho F, Dinis-Oliveira RJ, Silvestre R. Profiling of RNA degradation for estimation of post morterm interval. PLoS ONE. 2013;8:e56507.
Hynd MR, Lewohl JM, Scott HL, Dodd PR. Biochemical and molecular studies using human autopsy brain tissue. J Neurochem. 2003;85:543–62.
Vennemann M, Koppelkamm A. mRNA profiling in forensic genetics I: possibilities and limitations. Forensic Sci Int. 2010;203:71–5.
Zubakov D, Hanekamp E, Kokshoorn M, van IJcken W, Kayser M. Stable RNA markers for identification of blood and saliva stains revealed from whole genome expression analysis of time-wise degraded samples. Int J Legal Med. 2008;122:135–42.
Lee J, Hever A, Willhite D, Zlotnik A, Hevezi P. Effects of RNA degradation on gene expression analysis of human postmortem tissues. FASEB J. 2005;19:1356–8.
Gupta S, Halushka MK, Hilton GM, Arking DE. Postmortem cardiac tissue maintains gene expression profile even after late harvesting. BMC Genom. 2012;13:26.
Koppelkamm A, Vennemann B, Fracasso T, Lutz-Bonengel S, Schmidt U, Heinrich M. Validation of adequate endogenous reference genes for the normalisation of qPCR gene expression data in human post mortem tissue. Int J Legal Med. 2010;124:371–80.
Koppelkamm A, Vennemann B, Lutz-Bonengel S, Fracasso T, Vennemann M. RNA integrity in post-mortem samples: influencing parameters and implications on RT-qPCR assays. Int J Legal Med. 2011;125:573–80.
Durrenberger PF, Fernando S, Kashefi SN, Ferrer I, Hauw JJ, Seilhean D, et al. Effects of antemortem and postmortem variables on human brain mRNA quality: a BrainNet Europe study. J Neuropathol Exp Neurol. 2010;69:70–81.
Schulze-Osthoff K, Ferrari D, Los M, Wesselborg S, Peter ME. Apoptosis signaling by death receptors. Eur J Biochem. 1998;254:439–59.
Duprez L, Wirawan E, Berghe TV, Vandenabeele P. Major cell death pathways at a glance. Microbes Infect. 2009;11:1050–62.
Shalini S, Dorstyn L, Dawar S, Kumar S. Old, new and emerging functions of caspases. Cell Death Differ. 2015;22:526–39.
Acknowledgments
This work was supported by National Science Foundation (NSF) Grant HRD 1401075. The authors would like to thank Cuneyt Bademcioglu for his intellectual contribution and proofreading.
Author information
Authors and Affiliations
Corresponding author
Additional information
Gulnaz T. Javan and Ismail Can have contributed equally to this work.
Rights and permissions
About this article
Cite this article
Javan, G.T., Can, I., Finley, S.J. et al. The apoptotic thanatotranscriptome associated with the liver of cadavers. Forensic Sci Med Pathol 11, 509–516 (2015). https://doi.org/10.1007/s12024-015-9704-6
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12024-015-9704-6