Abstract
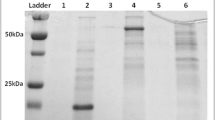
The xylanase gene (xynA) of Bacillus licheniformis 9945A was cloned and expressed in Escherichia coli BL21(DE3) using pET-22b(+) as an expression vector. The recombinant xylanase enzyme was purified by ammonium sulfate precipitation, followed by single-step immobilized metal ion affinity chromatography with a 57.58-fold purification having 138.2 U/mg specific activity and recovery of 70.08 %. Molecular weight of the purified xylanase, 23 kDa, was determined by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). The enzyme was stable for up to 70 °C with a broad pH range of 4–9 pH units. The enzyme activity was increased in the presence of metal ions especially Ca+2 and decreased in the presence of EDTA, indicating that the xylanase was a metalloenzyme. However, an addition of 1–4 % Tween 80, β-mercaptoethanol, and DTT resulted in the increase of enzyme activity by 51, 52, and 5 %, respectively. Organic solvents with a concentration of 10–40 % slightly decreased the enzyme activity. The xylanase enzyme possesses the ability of bioconversion of plant biomasses like wheat straw, rice straw, and sugarcane bagasse. Among the different tested biomasses, the highest saccharification percentage was observed with 1 % sugarcane bagasse after 72 h of incubation at 50 °C with 20 units of enzyme. The results suggest that recombinant xylanase can be used in the bioconversion of natural biomasses into simple sugars which could be further used for the production of biofuel.




Similar content being viewed by others
References
Gupta, N., Reddy, V. S., Maiti, S., & Ghosh, A. (2000). Cloning, expression, and sequence analysis of the gene encoding the alkali-stable, thermostable endoxylanase from alkalophilic, mesophilic Bacillus sp. strain NG-27. Applied and Environmental Microbiology, 66, 2631–2635.
Ninawe, S., Kapoor, M., & Kuhad, R. C. (2008). Purification and characterization of extracellular xylanase from Streptomyces cyaneus SN32. Bioresource Technology, 99, 1252–1258.
Ghangas, G. S., Hu, Y. J., & Wilson, D. (1989). Cloning of a Thermomonospora fusca xylanase gene and its expression in Escherichia coli and Streptomyces lividans. Journal of Bacteriology, 171, 2963–2969.
Loera Corral, O., Villasenor-Ortega, F., Guevara-Gonzalez, R., & Torres-Pacheco, I. (2006). Xylanases. Advances in Agricultural and Food Biotechnology, 2006, 305–322.
Tachaapaikoon, C., Kyu, K. L., & Ratanakhanokchai, K. (2006). Purification of xylanase from alkaliphilic Bacillus sp. K-8 by using corn husk column. Process Biochemistry, 41, 2441–2445.
Chidi, S. B., Godana, B., Ncube, I., Van Rensburg, E. J., Cronshaw, A., & Abotsi, E. K. (2008). Production, purification and characterization of celullase-free xylanase from Aspergillus terreus UL 4209. African Journal of Biotechnology, 7(21), 3939–3948.
Collins, T., Gerday, C., & Feller, G. (2005). Xylanases, xylanase families and extremophilic xylanases. FEMS Microbiology Reviews, 29, 3–23.
Pollet, A., Sansen, S., Raedschelders, G., Gebruers, K., Rabijns, A., Delcour, J. A., & Courtin, C. M. (2009). Identification of structural determinants for inhibition strength and specificity of wheat xylanase inhibitors TAXI‐IA and TAXI‐IIA. FEBS Journal, 276, 3916–3927.
Oakley, A. J., Heinrich, T., Thompson, C. A., & Wilce, M. C. (2003). Characterization of a family 11 xylanase from Bacillus subtillis B230 used for paper bleaching. Acta Crystallographica Section D: Biological Crystallography, 59, 627–636.
Beg, Q., Kapoor, M., Mahajan, L., & Hoondal, G. (2001). Microbial xylanases and their industrial applications: a review. Applied Microbiology and Biotechnology, 56, 326–338.
Subramaniyan, S., & Prema, P. (2002). Biotechnology of microbial xylanases: enzymology, molecular biology, and application. Critical Reviews in Biotechnology, 22, 33–64.
Kamble, R. D., & Jadhav, A. R. (2012). Isolation, purification, and characterization of xylanase produced by a new species of Bacillus in solid state fermentation. International Journal of Microbiology, 2012(2), 1–8.
Korona, B., Korona, D., & Bielecki, S. (2006). Efficient expression and secretion of two co-produced xylanases from Aspergillus niger in Pichia pastoris directed by their native signal peptides and the Saccharomyces cerevisiae α-mating factor. Enzyme and Microbial Technology, 39, 683–689.
Jamil, A., Nairn, S., Ahmad, S., & Ashraf, M. (2005). Production of industrially important enzymes using molecular approaches with special reference to xylanases and cellulases. Genetic Resources and Biotechnology, 2, 143.
Slapack, G. E., Russell, I., & Stewart, G. G. (1987). Thermophilic microbes in ethanol production.
Screenath, H. K., Jeffries, W. T. (2000). Bioresour Technol., 253–60.
Headon, D., & Walsh, G. (1994). The industrial production of enzymes. Biotechnology Advances, 12, 635–646.
Schoemaker, H. E., Mink, D., & Wubbolts, M. G. (2003). Dispelling the myths biocatalysis in industrial synthesis. Science, 299, 1694–1697.
Cazemier, A. E., Verdoes, J. C., Van Ooyen, A. J., & den Camp, H. J. O. (1999). Molecular and biochemical characterization of two xylanase-encoding genes from Cellulomonas pachnodae. Applied and Environmental Microbiology, 65, 4099–4107.
Srivastava, P., & Mukherjee, K. (2001). Cloning, characterization, and expression of xylanase gene from Bacillus lyticus in Escherichia coli and Bacillus subtilis. Preparative Biochemistry and Biotechnology, 31, 389–400.
Rey, M. W., Ramaiya, P., Nelson, B. A., Brody-Karpin, S. D., Zaretsky, E. J., Tang, M., & Clausen, I. G. (2004). Complete genome sequence of the industrial bacterium Bacillus licheniformis and comparisons with closely related Bacillus species. Genome Biology, 5, 77.
Kronstad, J., Schnepf, H., & Whiteley, H. (1983). Diversity of locations for Bacillus thuringiensis crystal protein genes. Journal of Bacteriology, 154, 419–428.
Cohen, S. N., Chang, A. C., & Hsu, L. (1972). Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proceedings of the National Academy of Sciences, 69, 2110–2114.
Bimboim, H., & Doly, J. (1979). A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Research, 7, 1513–1523.
Laemmeli, U. (1970). Cleavage of structural B proteins during the assembly of the head to bacteriophage T4. Nature, 227, 680–685.
Miller, G. (1959). Use of dinitrosalicylic acid reagent for determination reducing sugar. Analytical Chemistry, 31, 426–428.
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry, 72, 248–254.
de Moraes, L. M., Astolfi Filho, S., & Ulhoa, C. J. (1999). Purification and some properties of an α-amylase glucoamylase fusion protein from Saccharomyces cerevisiae. World Journal of Microbiology and Biotechnology, 15, 561–564.
Grayson, M., Graham-Rowe, D., Sanderson, K., Martin, M., & Lynd, L. R. (2011). Woods. Nature, 2011, 1–24.
Turner, P., Mamo, G., & Karlsson, E. N. (2007). Potential and utilization of thermophiles and thermostable enzymes in biorefining. Microbial Cell Factories, 6, 1–23.
Roayaei, M., & Galehdari, H. (2007). Cloning and expression of Thermus aquaticus DNA polymerase in Escherichia coli. Jundishapur Journal of Microbiology, 1, 1–5.
Gupta, N., Mehra, G., & Gupta, R. (2004). A glycerol-inducible thermostable lipase from Bacillus sp. medium optimization by a Plackett–Burman design and by response surface methodology. Canadian Journal of Microbiology, 50, 361–368.
Bhasin, A., Razdan, K., Gupta, N., & Sethi, N. (2014). Production and Purification of alkali stable xylanase from Bacillus sp. International Journal of Current Microbiology and Applied Science, 3, 365–377.
Meera, P., Wallner, M., & Otis, T. S. (2011). Molecular basis for the high THIP/gaboxadol sensitivity of extrasynaptic GABAA receptors. Journal of Neurophysiology, 106, 2057–2064.
Afzal, S., Saleem, M., Yasmin, R., Naz, M., & Imran, M. (2010). Pre and post cloning characterization of a β-1, 4-endoglucanase from Bacillus sp. Molecular Biology Reports, 37, 1717–1723.
Knob, A., Beitel, S. M., Fortkamp, D., Terrasan, C. R. F., & Almeida, A. F. d. (2013). Production, purification, and characterization of a major Penicillium glabrum xylanase using Brewer’s spent grain as substrate. BioMed research international, 2013.
Nakamura, S., Wakabayashi, K., Nakai, R., Aono, R., & Horikoshi, K. (1993). Purification and some properties of an alkaline xylanase from alkaliphilic Bacillus sp. strain 41M-1. Applied and Environmental Microbiology, 59, 2311–2316.
Chundakkadu Asha, P. (2011). Purification and biochemical characterization of xylanases from Bacillus pumilus and their potential for hydrolysis of polysaccharides. Fermentation Technology, 1, 1–8.
Akhavan Sepahy, A., Ghazi, S., & Akhavan Sepahy, M. (2011). Cost-effective production and optimization of alkaline xylanase by indigenous Bacillus mojavensis AG137 fermented on agricultural waste. Enzyme Research, 2011(2011), 1–9.
Huang, J., Wang, G., & Xiao, L. (2006). Cloning, sequencing and expression of the xylanase gene from a Bacillus subtilis strain B10 in Escherichia coli. Bioresource Technology, 97, 802–808.
Khajeh, K., Khezre-Barati, S., & Nemat-Gorgani, M. (2001). Proteolysis of mesophilic and thermophilic α-amylases. Applied Biochemistry and Biotechnology, 94, 97–109.
Gallardo, O., Diaz, P., & Pastor, F. J. (2004). Cloning and characterization of xylanase A from the strain Bacillus sp. BP-7: comparison with alkaline pI-low molecular weight xylanases of family 11. Current Microbiology, 48, 276–279.
Chivero, E. T., Mutukumira, A. N., & Zvauya, R. (2001). Partial purification and characterization of a xylanase enzyme produced by a micro-organism isolated from selected indigenous fruits of Zimbabwe. Food Chemistry, 72, 179–185.
Nair, S. G., Sindhu, R., & Shashidhar, S. (2008). Purification and biochemical characterization of two xylanases from Aspergillus sydowii SBS 45. Applied Biochemistry and Biotechnology, 149, 229–243.
Heck, J. X., de Barros Soares, L. H., Hertz, P. F., & Ayub, M. A. Z. (2006). Purification and properties of a xylanase produced by Bacillus circulans BL53 on solid-state cultivation. Biochemical Engineering Journal, 32, 179–184.
Bataillon, M., Cardinali, A. P. N., Castillon, N., & Duchiron, F. (2000). Purification and characterization of a moderately thermostable xylanase from Bacillus sp. strain SPS-0. Enzyme and Microbial Technology, 26, 187–192.
Yuan, X., Wang, J., Yao, H., & Venant, N. (2005). Separation and identification of endoxylanases from Bacillus subtilis and their actions on wheat bran insoluble dietary fibre. Process Biochemistry, 40, 2339–2343.
Khandeparker, R., Verma, P., & Deobagkar, D. (2011). A novel halotolerant xylanase from marine isolate Bacillus subtilis cho40: gene cloning and sequencing. New Biotechnology, 28, 814–821.
Faulet, B. M., Niamke, S., Gonnety, J. T., & Kouame, L. P. (2006). Purification and biochemical properties of a new thermostable xylanase from symbiotic fungus, Termitomyces sp. African Journal of Biotechnology, 5, 273–282.
Faulet, B. M., Niamké, S., Gonnety, J. T., & Kouamé, L. P. (2006). Purification and characterization of two thermostable cellulase-free xylanases from workers of the termite Macrotermes subhyalinus (Isoptera: Termitidae). International Journal of Tropical Insect Science, 26, 108.
Fialho, M., & Carmona, E. (2004). Purification and characterization of xylanases from Aspergillus giganteus. Folia Microbiologica, 49, 13–18.
Dutta, T., Sengupta, R., Sahoo, R., Sinha Ray, S., Bhattacharjee, A., & Ghosh, S. (2007). A novel cellulase free alkaliphilic xylanase from alkali tolerant Penicillium citrinum: production, purification and characterization. Letters in Applied Microbiology, 44, 206–211.
Do, T., Dam, T., & Quyen, D. (2009). Purification and biophysical characterization of xylanase from Aspergillus niger DSM 1957. Science Technology Journal of Agriculture and Rural Development, 6, 16–21.
Yang, Y., Zhang, W., Huang, J., Lin, L., Lian, H., Lu, Y., & Wang, S. (2010). Purification and characterization of an extracellular xylanase from Aspergillus niger C3486. African Journal of Microbiology Research, 4, 2249–2256.
Jeya, M., Thiagarajan, S., Lee, J.-K., & Gunasekaran, P. (2009). Cloning and expression of GH11 xylanase gene from Aspergillus fumigatus MKU1 in Pichia pastoris. Journal of Bioscience and Bioengineering, 108, 24–29.
Chapla, D., Divecha, J., Madamwar, D., & Shah, A. (2010). Utilization of agro-industrial waste for xylanase production by Aspergillus foetidus MTCC 4898 under solid state fermentation and its application in saccharification. Biochemical Engineering Journal, 49, 361–369.
Coughlan, M. (1991). Towards an understanding of the mechanism of action of main chain-hydrolyzing xylanases. Progress in Biotechnology, 7, 111–139.
Dhabhai, R., Jain, A., & Chaurasia, S. P. (2012). Production of fermentable sugars by dilute acid pretreatment and enzymatic saccharification of three different lignocellulosic materials. International Journal of Chemistry Technology Research, 4, 1497–1502.
Mamo, G., Thunnissen, M., Hatti-Kaul, R., & Mattiasson, B. (2009). An alkaline active xylanase: insights into mechanisms of high pH catalytic adaptation. Biochimie, 91, 1187–1196.
Liang, Y., Feng, Z., Yesuf, J., & Blackburn, J. W. (2010). Optimization of growth medium and enzyme assay conditions for crude cellulase produced by a novel thermophilic and cellulolytic bacterium, Anoxybacillus sp. 527. Applied Biochemistry and Biotechnology, 160, 1841–1852.
Nagar, S., Gupta, V. K., Kumar, D., Kumar, L., & Kuhad, R. C. (2010). Production and optimization of cellulase-free, alkali-stable xylanase by Bacillus pumilus SV-85S in submerged fermentation. Journal of Industrial Microbiology & Biotechnology, 37, 71–83.
Acknowledgments
Sincere gratitude is expressed to the Higher Education Commission (HEC), Islamabad, Pakistan, for funding this project as PhD research work for Ms. Asma Zafar.
Author information
Authors and Affiliations
Corresponding author
Additional information
Asma Zafar and Muhammad Nauman Aftab contributed equally to this work.
Rights and permissions
About this article
Cite this article
Zafar, A., Aftab, M.N., Din, Z.u. et al. Cloning, Expression, and Purification of Xylanase Gene from Bacillus licheniformis for Use in Saccharification of Plant Biomass. Appl Biochem Biotechnol 178, 294–311 (2016). https://doi.org/10.1007/s12010-015-1872-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-015-1872-z