Abstract
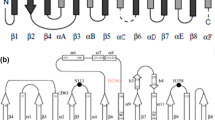
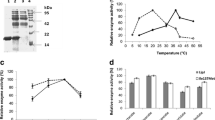
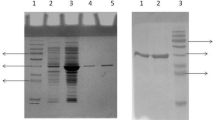
Lipases from Bacillus thermocatenulatus are a member of superfamily of α/β hydrolase, but there are structural differences between them. In this work, we focused on the α5 helix of B. thermocatenulatus lipase (BTL2) which is absent in canonical α/β hydrolase fold. In silico study showed that the α5 helix is a region that causes disorder in BTL2 protein. So, the α5 helix (residues 131 to 150) has been deleted from the B. thermocatenulatus lipase gene (BTL2) and the remain (Δα5-BTL2) has been expressed in Pichia pastoris yeast. The α5 deletion results in increase of enzyme-specific activity in the presence of tributyrin, tricaproin, tricaprylin, tricaprin, trilaurin, and olive oil (C18) substrates by 1.4-, 1.7-, 2.0-, 1.2-, 1.75-, and 1.95-fold, respectively. Also, deletion leads to increase in enzyme activity in different temperatures and pHs, whereas it did not significantly affect on enzyme activity in the presence of organic solvents, metal ions, and detergents.









Similar content being viewed by others
Abbreviations
- BMMH:
-
Buffered minimal medium containing histidine
- YPG:
-
Yeast extract peptone dextrose medium
References
Khare, S. K., Nabetani, H., & Nakajima, M. (2000). Lipase catalyzed interesterification reaction and their industrial applications. Indian Food Industry, 19, 29–35.
Carriere, F., Thirstrup, K., Hjorth, S., & Boel, E. (1994). Cloning of the classical guinea pig pancreatic lipase and comparison with the lipase related protein. FEBS Letters, 338, 63–68.
Kademi, A., Danielle, L., Again, H. (2005). Lipases. In: Enzyme technology (pp. 297–318). New Delhi, India: Asiatech.
Sharma, R., Chisti, Y., & Banerjee, U. C. (2001). Production, purification, characterization and application of lipases. Biotechnology Advances, 19, 627–662.
Hasan, F., Ali Shah, A., & Hameed, A. (2006). Industrial application of microbial lipases. Enzyme and Microbial Technology, 39, 235–251.
Salis, A., Meloni, M., Ligas, S., Casula, M. F., Monduzzi, M., Solinas, V., & Dumitriu, E. (2005). Physical and chemical adsorption of Mucor javanicus lipase on SBA-15 mesoporous silica. Synthesis, structural characterization, and activity performance. Langmuir, 21(12), 5511–5516.
Nazina, T. N., Tourova, T. P., Poltaraus, A. B., Novikova, E. V., Grigoryan, A. A., Ivanova, A. E., Lysenko, A. M., Petrunyaka, V. V., Osipov, G. A., Belyaev, S. S., & Ivanov, M. V. (2001). Taxonomic study of aerobic thermophilic bacilli: descriptions of Geobacillus subterraneus gen. nov., sp. nov. and Geobacillus uzenensis sp. nov. from petroleum reservoirs and transfer of Bacillus stearothermophilus, Bacillus thermocatenulatus, Bacillus thermoleovorans, Bacillus kaustophilus, Bacillus thermoglucosidasius and Bacillus thermodenitrificans to Geobacillus as the new combinations. International Journal of Systematic and Evolutionary Microbiology, 51, 433–446.
Tyndall, J., Sinchaikul, S., Fothergill-Gilmore, L., Taylor, P., & Walkinshaw, M. (2002). Crystal structure of thermostable lipase from Bacillus stearothermophilus P1. Journal of Molecular Biology, 323, 859–869.
Carrasco-Lopez, C., Godoy, C., Rivas, B., Lorente, G., Palomo, J., Guisan, J., Lafuente, R., Ripoll, M., & Hermosa, J. (2009). Activation of bacterial thermoalkalophilic lipases is spurred by dramatic structural rearrangements. Journal of Biological Chemistry, 284, 4365–4372.
Nardini, M., & Dijkstra, B. W. (1999). α/β hydrolase fold enzymes: the family keeps growing. Current Opinion in Structural Biology, 9, 732–737.
Joseph, B., Ramteke, P. W., & Thomas, G. (2008). Cold active microbial lipases: some hot issues and recent developments. Biotechnology Advances, 26(5), 457–470.
Jaeger, K. E., Ransac, S., Dijkstra, B. W., Colson, C., Heuvel, M. V., & Misset, O. (1994). Bacterial lipases. FEMS Microbiology Reviews, 15, 29–63.
Pouderoyen, G. V., Eggert, T., Jaeger, K., & Dijkstra, B. W. (2001). The crystal structure of Bacillus subtilis lipase: a minimal α/β hydrolase fold enzyme. Journal of Molecular Biology, 309, 215–226.
Angkawidjaja, C., & Kanaya, S. (2006). Family I.3 lipase: bacterial lipases secreted by the type I secretion system. Cellular and Molecular Life Sciences, 63, 2804–2817.
Laskowski, R. A., Macarthur, M. W., Moss, D. S., & Thornton, J. M. (1993). PROCHECK: a program to check the stereochemical quality of protein structures. Journal of Applied Crystallography, 26, 283–291.
Colovos, C., & Yeates, T. O. (1993). Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sciences, 2, 1511–1519.
Benkert, P., Tosatto, S. C. E., & Schomburg, D. (2008). QMEAN: a comprehensive scoring function for model quality assessment. Proteins: Structure, Function, and Bioinformatics, 71, 261–277.
Wiederstein, M., & Sippl, M. J. (2007). Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Research, 35, W407–W410.
Humphrey, W., Dalke, A., & Sculten, K. (1996). VMD: visual molecular dynamics. Journal of Molecular Graphics, 14, 33–38.
Karkhane, A. A., Yakhchali, B., Rastgar Jazii, F., & Bambai, B. (2009). The effect of substitution of Phe181 and Phe 182 with Ala on activity, substrate specificity and stabilization of substrate at the active site of Bacillus thermocatenulatus lipase. Journal of Molecular Catalysis B: Enzymatic, 61, 162–167.
Eswar, N., Marti-Renom, M. A., Webb, B., Madhusudhan, M., Eramian, D., Shen, M., Pieper, U., Sali, A. (2006). Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinformatics. John Wiley & Sons, Inc.
Hosseini, M., Karkhane, A. A., Yakhchali, B., Shamsara, M., Aminzadeh, S., Morshedi, D., Haghbeen, K., Torktaz, I., Karimi, E., & Safari, Z. (2012). In silico and experimental characterization of chimeric Bacillus thermocatenulatus lipase with the complete conserved pentapeptide of Candida rugosa lipase. Applied Biochemistry and Biotechnology, 169, 773–785.
Warrens, A. N., Jones, M. D., & Lechler, R. I. (1997). Splicing by overlap extension by PCR using asymmetric amplification: an improved technique for generation of hybrid proteins of immunological interest. Gene, 186, 29–35.
Karkhane, A. A., Yakhchali, B., Rastgar Jazii, F., Hemmat, J., Shariati, P., Khodabandeh, M., & Zomorodipoor, A. (2012). Periplasmic expression of Bacillus thermocatenulatus lipase in Escherichia coli in presence of different signal sequences. Iranian Journal of Biotechnology, 10, 255–262.
Quyen, D. T., Schmidt-Dannert, C., & Schmid, R. D. (2003). High-level expression of a lipase from Bacillus thermocatenulatus BTL2 in Pichia pastoris and some properties of the recombinant lipase. Protein Expression and Purification, 28, 102–110.
Laemmli, U. K. (1971). Nature, 227, 680.
Rastgar Jazii, F., Karkhane, A. A., Yakhchali, B., Fatemi, S. A., & Deezagi, A. A. (2007). Simplified purification procedure for recombinant human granulocyte macrophage colony stimulating factor from periplasmic space of Escherichia coli. Journal of Chromatography, 856, 214–221.
Benkert, P., Biasini, M., & Schwede, T. (2011). Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics, 10, 1093.
Abdul Rahman, M. Z., Salleh, A. B., Noor Zaliha, R., Abdul Rahman, R., Abdul Rahman, M. B., Basri, B., & Chor, L. (2012). Unlocking the mystery behind the activation phenomenon of T1 lipase: a molecular dynamics simulations approach. Protein Sciences, 21, 1210–1221.
Wahab, R., Basri, M., Rahman, M., Rahman, R., Salleh, A., & Chor, L. (2012). Engineering catalytic efficiency of thermophilic lipase from Geobacillus zalihae by hydrophobic residue mutation near the catalytic pocket. Advances in Bioscience and Biotechnology, 3, 158–167.
Ghori, M. I., Iqbal, M. J., & Hameed, A. (2011). Characterization of a novel lipase from Bacillus sp. isolated from tannery wastes. Brazilian Journal of Microbiology, 42, 22–29.
Karimi, E., Karkhane, A. A., Yakhchali, B., Shamsara, M., Aminzadeh, S., Torktaz, I., Hosseini, M., & Safari, Z. (2014). Study of the effect of F17A mutation on characteristics of Bacillus thermocatenulatus lipase expressed in Pichia pastoris using in silico and experimental methods. Biotechnology and Applied Biochemistry, 61, 264.
Novototskaya-Vlasova, K. A., Petrovskaya, L. E., Rivkina, E. M., Dolgikh, D. A., & Kirpichnikov, M. P. (2013). Characterization of a cold-active lipase from Psychrobacter cryohalolentis K5T and its deletion mutants. Biochemistry (Moscow), 78(4), 385–394.
Acknowledgments
We would like to express our appreciation and thanks to Mrs. F. Rahimi and Dr. P. Farrokh for their help on the accomplishment of this study. We are also grateful to Mr. H. Jamali for the language editing of our manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Goodarzi, N., Karkhane, A.A., Mirlohi, A. et al. Protein Engineering of Bacillus thermocatenulatus Lipase via Deletion of the α5 Helix. Appl Biochem Biotechnol 174, 339–351 (2014). https://doi.org/10.1007/s12010-014-1063-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-014-1063-3