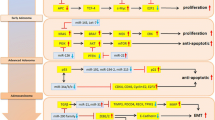
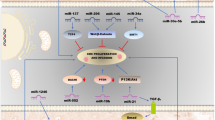
Abstract
MicroRNAs (or miRNAs) are small transcripts of 20–24 nucleotides that have emerged as important regulators of gene expression in cancer cells. Overexpression and silencing of specific miRNAs has been linked to the stepwise disease progression during the normal adenoma-cancer sequence in colorectal cancer (CRC). Given their cancer-specific pattern of expression, remarkable stability, and presence in blood and other body fluids, miRNAs are considered to be highly promising cancer biomarkers. Although the use of miRNA biomarkers for CRC is still in its infancy, accumulating evidence firmly supports the existence of unique “miRNA signatures” that can not only facilitate earlier detection of the tumor, but can also assist in predicting disease recurrence and therapeutic outcome to currently available treatment regimens. There is a great hope and promise that some of these miRNA biomarkers will eventually be validated in large-scale clinical trials, and if successful, it will open new avenues for the earlier diagnosis and personalization of therapy for CRC. In this review, we summarize the current state of literature on the potential diagnostic, prognostic, and predictive potential of miRNA biomarkers in CRC.
Similar content being viewed by others
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300.
Meissner HI, Breen N, Klabunde CN, Vernon SW. Patterns of colorectal cancer screening uptake among men and women in the United States. Cancer Epidem Biomar. 2006;15:389–94.
Kim DH, Pickhardt PJ, Taylor AJ, et al. CT colonography versus colonoscopy for the detection of advanced neoplasia. N Engl J Med. 2007;357:1403–12.
Levin B, Lieberman DA, McFarland B, et al. Screening and surveillance for the early detection of colorectal cancer and adenomatous polyps, 2008: a joint guideline from the American Cancer Society, the US Multi-Society Task Force on Colorectal Cancer, and the American College of Radiology. CA Cancer J Clin. 2008;58:130–60.
Graser A, Stieber P, Nagel D, et al. Comparison of CT colonography, colonoscopy, sigmoidoscopy and faecal occult blood tests for the detection of advanced adenoma in an average risk population. Gut. 2009;58:241–8.
Mandel JS, Bond JH, Church TR, et al. Reducing mortality from colorectal cancer by screening for fecal occult blood. Minnesota colon cancer control study. N Engl J Med. 1993;328:1365–71.
Ouyang DL, Chen JJ, Getzenberg RH, Schoen RE. Noninvasive testing for colorectal cancer: a review. Am J Gastroenterol. 2005;100:1393–403.
Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–5.
Zeng Y, Yi R, Cullen BR. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc Natl Acad Sci USA. 2003;100:9779–84.
Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–8.
Calin GA, Croce CM. MicroRNA signatures in human cancers. Nature reviews. Cancer. 2006;6:857–66.
••Schetter AJ, Leung SY, Sohn JJ et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA 2008; 299: 425–436. This article was the first one to perform microarray-based profiling for miRNAs in cancer tissues, and to identify the prognostic and therapeutic potential of miR-21, which was later confirmed in multiple reports.
Hui AB, Shi W, Boutros PC, et al. Robust global micro-RNA profiling with formalin-fixed paraffin-embedded breast cancer tissues. Lab Invest. 2009;89:597–606.
Weizman AV, Nguyen GC. Colon cancer screening in 2010: an up-date. Minerva Gastroenterol Dietol. 2010;56:181–8.
Imperiale TF, Ransohoff DF, Itzkowitz SH, et al. Fecal DNA versus fecal occult blood for colorectal-cancer screening in an average-risk population. N Engl J Med. 2004;351:2704–14.
Zou H, Taylor WR, Harrington JJ, et al. High detection rates of colorectal neoplasia by stool DNA testing with a novel digital melt curve assay. Gastroenterology. 2009;136:459–70.
Schmitz KJ, Hey S, Schinwald A, et al. Differential expression of microRNA 181b and microRNA 21 in hyperplastic polyps and sessile serrated adenomas of the colon. Virchows Arch. 2009;455:49–54.
Yamamichi N, Shimomura R, Inada K, et al. Locked nucleic acid in situ hybridization analysis of miR-21 expression during colorectal cancer development. Clin Cancer Res. 2009;15:4009–16.
Fassan M, Pizzi M, Giacomelli L, et al. PDCD4 nuclear loss inversely correlates with miR-21 levels in colon carcinogenesis. Virchows Arch. 2011;458:413–9.
Chang KH, Miller N, Kheirelseid EA, et al. MicroRNA-21 and PDCD4 expression in colorectal cancer. Eur J Surg Oncol. 2011;37:597–603.
Diosdado B, van de Wiel MA. Terhaar Sive Droste JS et al. MiR-17-92 cluster is associated with 13q gain and c-myc expression during colorectal adenoma to adenocarcinoma progression. Br J Cancer. 2009;101:707–14.
Nagel R, le Sage C, Diosdado B, et al. Regulation of the adenomatous polyposis coli gene by the miR-135 family in colorectal cancer. Cancer Res. 2008;68:5795–802.
Akao Y, Nakagawa Y, Hirata I, et al. Role of anti-oncomirs miR-143 and -145 in human colorectal tumors. Cancer Gene Ther. 2010;17:398–408.
Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–61.
Wang CJ, Zhou ZG, Wang L, et al. Clinicopathological significance of microRNA-31, -143 and -145 expression in colorectal cancer. Dis Markers. 2009;26:27–34.
Bandres E, Cubedo E, Agirre X, et al. Identification by Real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues. Mol Cancer. 2006;5:29.
Slaby O, Svoboda M, Fabian P, et al. Altered expression of miR-21, miR-31, miR-143 and miR-145 is related to clinicopathologic features of colorectal cancer. Oncology. 2007;72:397–402.
Balaguer F, Moreira L, Lozano JJ, et al. Colorectal Cancers with Microsatellite Instability Display Unique miRNA Profiles. Clin Cancer Res. 2011;17:6239–49.
Lawrie CH, Gal S, Dunlop HM, et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. 2008;141:672–5.
Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513–8.
Lodes MJ, Caraballo M, Suciu D, et al. Detection of cancer with serum miRNAs on an oligonucleotide microarray. PLoS One. 2009;4:e6229.
Chen X, Ba Y, Ma L, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006.
••Ng EK, Chong WW, Jin H et al. Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut 2009; 58: 1375-1381. This was a seminal study that demonstrated the usefulness of plasma miRNAs as diagnostic and prognostic biomarkers for colorectal cancer. In addition, the panel of miRNA biomarkers could successfully delineate CRC from gastric cancer, IBD, and normal patients.
Huang Z, Huang D, Ni S, et al. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer. 2010;127:118–26.
Wang LG, Gu J. Serum microRNA-29a is a promising novel marker for early detection of colorectal liver metastasis. Cancer epidemiology 2011.
Kroh EM, Parkin RK, Mitchell PS, Tewari M. Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR). Methods. 2010;50:298–301.
Ahlquist DA. Molecular detection of colorectal neoplasia. Gastroenterology. 2010;138:2127–39.
•Link A, Balaguer F, Shen Y et al. Fecal MicroRNAs as novel biomarkers for colon cancer screening. Cancer Epidemiol Biomarkers Prev 2010; 19: 1766-1774. This was the first report that demonstrated the feasibility of fecal miRNA extraction, and performed an array-based profiling to identify fecal miRNAs that can identify colorectal neoplasia.
Ahmed FE, Jeffries CD, Vos PW, et al. Diagnostic microRNA markers for screening sporadic human colon cancer and active ulcerative colitis in stool and tissue. Cancer Genomics Proteomics. 2009;6:281–95.
Koga Y, Yasunaga M, Takahashi A, et al. MicroRNA expression profiling of exfoliated colonocytes isolated from feces for colorectal cancer screening. Cancer Prev Res (Phila). 2010;3:1435–42.
Wu CW, Ng SS, Dong YJ et al. Detection of miR-92a and miR-21 in stool samples as potential screening biomarkers for colorectal cancer and polyps. Gut.
Kalimutho M, Del Vecchio Blanco G, Di Cecilia S et al. Differential expression of miR-144* as a novel fecal-based diagnostic marker for colorectal cancer. J Gastroenterol 2011.
Kulda V, Pesta M, Topolcan O, et al. Relevance of miR-21 and miR-143 expression in tissue samples of colorectal carcinoma and its liver metastases. Cancer Genet Cytogenet. 2010;200:154–60.
Shibuya H, Iinuma H, Shimada R, et al. Clinicopathological and prognostic value of microRNA-21 and microRNA-155 in colorectal cancer. Oncology. 2010;79:313–20.
Xi Y, Formentini A, Chien M, et al. Prognostic Values of microRNAs in Colorectal Cancer. Biomark Insights. 2006;2:113–21.
Akcakaya P, Ekelund S, Kolosenko I et al. miR-185 and miR-133b deregulation is associated with overall survival and metastasis in colorectal cancer. Int J Oncol 39: 311-318.
Motoyama K, Inoue H, Takatsuno Y, et al. Over- and under-expressed microRNAs in human colorectal cancer. Int J Oncol. 2009;34:1069–75.
Diaz R, Silva J, Garcia JM, et al. Deregulated expression of miR-106a predicts survival in human colon cancer patients. Genes Chromosomes Cancer. 2008;47:794–802.
Wu Z, Zheng S, Li Z, et al. E2F1 suppresses Wnt/beta-catenin activity through transactivation of beta-catenin interacting protein ICAT. Oncogene. 2011;30:3979–84.
Hu G, Chen D, Li X et al. miR-133b regulates the MET proto-oncogene and inhibits the growth of colorectal cancer cells in vitro and in vivo. Cancer Biol Ther 10: 190-197.
Ribic CM, Sargent DJ, Moore MJ, et al. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N Engl J Med. 2003;349:247–57.
Schepeler T, Reinert JT, Ostenfeld MS, et al. Diagnostic and prognostic microRNAs in stage II colon cancer. Cancer Res. 2008;68:6416–24.
Schetter AJ, Nguyen GH, Bowman ED, et al. Association of inflammation-related and microRNA gene expression with cancer-specific mortality of colon adenocarcinoma. Clin Cancer Res. 2009;15:5878–87.
Nielsen BS, Jorgensen S, Fog JU, et al. High levels of microRNA-21 in the stroma of colorectal cancers predict short disease-free survival in stage II colon cancer patients. Clin Exp Metastasis. 2011;28:27–38.
Labianca R, Nordlinger B, Beretta GD, et al. Primary colon cancer: ESMO Clinical Practice Guidelines for diagnosis, adjuvant treatment and follow-up. Ann Oncol. 2010;21 Suppl 5:v70–7.
Cheng H, Zhang L, Cogdell DE, et al. Circulating plasma MiR-141 is a novel biomarker for metastatic colon cancer and predicts poor prognosis. PLoS One. 2011;6:e17745.
Katoh Y, Katoh M. Hedgehog signaling, epithelial-to-mesenchymal transition and miRNA (review). Int J Mol Med. 2008;22:271–5.
Nakajima G, Hayashi K, Xi Y, et al. Non-coding MicroRNAs hsa-let-7g and hsa-miR-181b are Associated with Chemoresponse to S-1 in Colon Cancer. Cancer Genomics Proteomics. 2006;3:317–24.
Svoboda M. Izakovicova Holla L, Sefr R et al. Micro-RNAs miR125b and miR137 are frequently upregulated in response to capecitabine chemoradiotherapy of rectal cancer. Int J Oncol. 2008;33:541–7.
Drebber U, Lay M, Wedemeyer I, et al. Altered levels of the onco-microRNA 21 and the tumor-supressor microRNAs 143 and 145 in advanced rectal cancer indicate successful neoadjuvant chemoradiotherapy. Int J Oncol. 2011;39:409–15.
Pu XX, Huang GL, Guo HQ, et al. Circulating miR-221 directly amplified from plasma is a potential diagnostic and prognostic marker of colorectal cancer and is correlated with p53 expression. J Gastroenterol Hepatol. 2010;25:1674–80.
Acknowledgment
The authors are highly grateful to Margaret Hinshelwood, PhD for her suggestions and skillful editing of the manuscript.
Disclosure
No potential conflicts of interest relevant to this article were reported.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Toiyama, Y., Goel, A. The Diagnostic, Prognostic, and Predictive Potential of MicroRNA Biomarkers in Colorectal Cancer. Curr Colorectal Cancer Rep 8, 22–31 (2012). https://doi.org/10.1007/s11888-011-0110-5
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11888-011-0110-5