Abstract
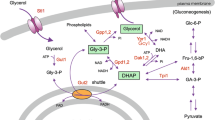
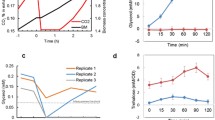
In order to maintain its turgor pressure at a desired homeostatic level, budding yeast, Saccharomyces cerevisiae responds to the external variation of the osmotic pressure by varying its internal osmotic pressure through regulation of synthesis and transport of the intracellular glycerol. Hog1PP (dually phosphorylated Hog1), a final effector in the signalling pathway of the hyper osmotic stress, regulates the glycerol synthesis both at transcriptional and non-transcriptional stages. It is known that for a step-change in salt concentration leading to moderate osmotic shock, Hog1PP activity shows a transient response before it returns to the vicinity of pre-stimulus level. It is believed that an integrating process in a negative feedback loop can be a design strategy to yield such an adaptive response. Several negative feedback loops have been identified in the osmoadaptation system in yeast. However, the precise location of the integrating process in the osmoadaptation system which includes signalling, gene regulation, metabolism and biophysical modules is unclear. To address this issue, we developed a reduced model which captures various experimental observations of the osmoadaptation behaviour of wild type and mutant strains. Dynamic simulations and steady state analysis suggested that known information about the osmoadaptation system of budding yeast does not necessarily give a perfect integrating process through the known feedback loops of Hog1PP. On the other hand, regulation of glycerol synthesising enzyme degradation can result in a near integrating process leading to a near-perfect adaptation.










Similar content being viewed by others
References
Adrover MÀ, Zi Z, Duch A, Schaber J, González-Novo A, Jimenez J, Nadal-Ribelles M, Clotet J, Klipp E, Posas F (2011) Time-dependent quantitative multicomponent control of the G 1-S network by the stress-activated protein kinase Hog1 upon osmostress. Sci Signal 4(192):ra63
Beese SE, Negishi T, Levin DE (2009) Identification of positive regulators of the yeast Fps1 glycerol channel. PLoS Genet 5(11):e1000738
Freire P, Vinod PK, Novak B (2012) Interplay of transcriptional and proteolytic regulation in driving robust cell cycle progression. Mol BioSyst 8:863–870
Hohmann S (2002) Osmotic stress signaling and osmoadaptation in yeasts. Microbiol Mol Biol Rev 66:300–372
Jain NK, Roy I (2010) Trehalose and protein stability. Curr Protoc Protein Sci 59:4.9.1–4.9.12
Judy C (1995) Regulation of protein degradation. Plant Cell 7:845–857
Klipp E, Nordlander B, Krüger R, Gennemark P, Hohmann S (2005) Integrative model of the response of yeast to osmotic shock. Nat Biotechnol 23:975–982
Macia J, Regot S, Peeters T, Conde N, Solé R, Posas F (2009) Dynamic signaling in the Hog1 MAPK pathway relies on high basal signal transduction. Sci Signal 2(63):ra13
Marrocco K, Bergdoll M, Achard P, Criqui M-C, Genschik P (2010) Selective proteolysis sets the tempo of the cell cycle. Curr Opin Plant Biol 13:631–639
Mettetal JT, Muzzey D, Gómez-Uribe C, Van Oudenaarden A (2008) The frequency dependence of osmo-adaptation in Saccharomyces cerevisiae. Science 319:482–484
Miermont A, Waharteb F, McCleand MN, Bottania S, Léone S, Hersena P (2013) Severe osmotic compression triggers a slowdown of intracellular signaling, which can be explained by molecular crowding. Proc Natl Acad Sci USA 110:5725–5730
Miller C, Schwalb B, Maier K, Schulz D, Dümcke S, Zacher B, Mayer A, Sydow J, Marcinowski L, Dölken L et al (2011) Dynamic transcriptome analysis measures rates of mRNA synthesis and decay in yeast. Mol Syst Biol 7:458
Muzzey D, Gómez-Uribe CA, Mettetal JT, van Oudenaarden A (2009) A systems-level analysis of perfect adaptation in yeast osmoregulation. Cell 138:160–171
Norbeck J, Blomberg A (1997) Metabolic and regulatory changes associated with growth of Saccharomyces cerevisiae in 1.4 M NaCl. Evidence for osmotic induction of glycerol dissimilation via the dihydroxyacetone pathway. J Biol Chem 272:5544–5554
Parmar JH, Bhartiya S, Venkatesh KV (2009) A model-based study delineating the roles of the two signaling branches of Saccharomyces cerevisiae, Sho1 and Sln1, during adaptation to osmotic stress. Phys Biol 6:036019
Petelenz-Kurdziel E, Eriksson E, Smedh M, Beck C, Hohmann S, Goksör M (2011) Quantification of cell volume changes upon hyperosmotic stress in Saccharomyces cerevisiae. Integr Biol 3:1120–1126
Petelenz-Kurdziel E, Kuehn C, Nordlander B, Klein D, Hong K–K, Jacobson T, Dahl P, Schaber J, Nielsen J, Hohmann S et al (2013) Quantitative analysis of glycerol accumulation, glycolysis and growth under hyper osmotic stress. PLoS Comput Biol 9:e1003084
Rep M, Reiser V, Gartner U, Thevelein JM, Hohmann S, Ammerer G, Ruis H (1999) Osmotic stress-induced gene expression in Saccharomyces cerevisiae requires Msn1p and the novel nuclear factor Hot1p. Mol Cell Biol 19:5474–5485
Romero-Santacreu L, Moreno J, Pérez-Ortín JE, Alepuz P (2009) Specific and global regulation of mRNA stability during osmotic stress in Saccharomyces cerevisiae. RNA 15:1110–1120
Schaber J, Baltanas R, Bush A, Klipp E, Colman-Lerner A (2012) Modelling reveals novel roles of two parallel signalling pathways and homeostatic feedbacks in yeast. Mol Syst Biol 8:622
Street TO, Bolen DW, Rose GD (2006) A molecular mechanism for osmolyte-induced protein stability. Proc Natl Acad Sci 103:13997–14002
Tamás MJ, Luyten K, Sutherland FCW, Hernandez A, Albertyn J, Valadi H, Li H, Prior BA, Kilian SG, Ramos J et al (1999) Fps1p controls the accumulation and release of the compatible solute glycerol in yeast osmoregulation. Mol Microbiol 31:1087–1104
Van Wuytswinkel O, Reiser V, Siderius M, Kelders MC, Ammerer G, Ruis H, Mager WH (2000) Response of Saccharomyces cerevisiae to severe osmotic stress: evidence for a novel activation mechanism of the HOG MAP kinase pathway. Mol Microbiol 37:382–397
Westfall PJ, Patterson JC, Chen RE, Thorner J (2008) Stress resistance and signal fidelity independent of nuclear MAPK function. Proc Natl Acad Sci USA 34:12212–12217
Yi T-M, Huang Y, Simon MI, Doyle J (2000) Robust perfect adaptation in bacterial chemotaxis through integral feedback control. Proc Natl Acad Sci USA 97:4649–4653
You T, Ingram P, Jacobsen MD, Cook E, McDonagh A, Thorne T, Lenardon MD, De Moura AP, Romano MC, Thiel M et al (2012) A systems biology analysis of long and short-term memories of osmotic stress adaptation in fungi. BMC Res Notes 5:258
Acknowledgments
A.K.P. acknowledges Parmar Jignesh for useful discussions. K.V.V. acknowledges funds from Department of Science and Technology (DST), India.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Patel, A.K., Bhartiya, S. & Venkatesh, K.V. Analysis of osmoadaptation system in budding yeast suggests that regulated degradation of glycerol synthesis enzyme is key to near-perfect adaptation. Syst Synth Biol 8, 141–154 (2014). https://doi.org/10.1007/s11693-013-9126-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11693-013-9126-2