Abstract
Mouse Zinc finger and SCAN domain containing 4 (Zscan4) is encoded in multiple copies of Zscan4 genes, which are expressed in late two-cell stage preimplantation embryos and in 1–5% of the embryonic stem (ES) cell population at a given time. Due to the highly identical nucleotide sequences of multiple copies of Zscan4 paralogs and pseudogenes in the mouse Zscan4 genomic cluster, previous analyses have been done using exogenous transgenes under the regulation of Zscan4c promoter. In this manuscript, we generated knock-in mouse ES cell lines and mouse lines, in which the expression of endogenous Zscan4c, one of the Zscan4 genes, can be specifically monitored with a green fluorescent protein variant, Emerald. Interestingly, we found that only ∼30% of Zscan4-immunopositive ES cells were Emerald positive, suggesting that even when the Zscan4 locus is active, not all Zscan4 genes are expressed synchronously. We also carried out mass spectrometry of protein complexes associated with endogenous Zscan4 proteins. Taken together, our genetic engineering at an endogenous Zscan4c gene provides the first clue for the expression and function of each gene copy of Zscan4 locus in a physiological context.
Similar content being viewed by others
Introduction
Zinc finger and SCAN domain containing 4 (Zscan4) genes were originally identified as genes that are specifically expressed during zygotic genome activation (ZGA) in the late two-cell stage of mouse preimplantation embryos (Falco et al. 2007). The mouse genome encodes nine members of the Zscan4 paralogs (Zscan4a, Zscan4b, Zscan4c, Zscan4d, Zscan4e, and Zscan4f and three pseudogenes Zscan4-ps1, Zscan4-ps2, and Zscan4-ps3). In the human genome, there is only one copy of Zscan4 gene (Falco et al. 2007). Among the mouse Zscan4 genes, Zscan4c, Zscan4d, and Zscan4f encode a full-length 506-aa protein, whereas Zscan4a, Zscan4b, and Zscan4e encode truncated proteins (360 amino acids (aa), 195 aa, and 195 aa, respectively) (Falco et al. 2007). In two-cell stage embryos, the knockdown of Zscan4 by small interfering RNA (siRNA) leads to a delay of progression from the two-cell to four-cell stage and, consequently, implantation failure (Falco et al. 2007). In mouse embryonic stem (ES) cells, the expression of Zscan4 is transient and reversible with infrequent transcriptional activation in only 1–5% of the cell population at a given time point (Falco et al. 2007) (Zalzman et al. 2010). A burst of Zscan4 transcription (Z4 events) is accompanied by biological events including transient expression of other ZGA-specific genes (Amano et al. 2013; Akiyama et al. 2015) rapid derepression and rerepression of heterochromatin regions (Akiyama et al. 2015), rapid telomere extension (Zalzman et al. 2010), and blockage of global translation (Hung et al. 2013). Additionally, Zscan4 has also been shown to enhance the efficiency of generating mouse-induced pluripotent stem (iPS) cells and their quality (Hirata et al. 2012; Jiang et al. 2013). These data suggest that Zscan4 plays diverse biological roles during Z4 events of ES cells and in two-cell stage preimplantation embryos.
In the previous studies, Z4 events were mostly identified in ES cells with a reporter transgene, in which the fluorescent reporter expression is under an artificial Zscan4c promoter region (Zalzman et al. 2010; Akiyama et al. 2015). However, a potential issue that has yet to be clarified is whether the minimum 3.6-kb genomic fragment of the putative Zscan4c promoter region mirrors the bona fide expression pattern of the endogenous Zscan4 locus due to random integration in the genome, copy number effect, and any missing cis-regulatory element. In addition, due to limited material available for biochemical examination, it has been elusive which member of the endogenous Zscan4 proteins act for Z4 events in ES cells and for preimplantation embryo development, although previous RNA-sequencing analyses suggest that all the members of Zscan4 messenger RNA (mRNA) are expressed (Akiyama et al. 2015), albeit Zscan4c is expressed predominantly in ES cells, and Zscan4d is expressed predominantly in two-cell stage embryos (Falco et al. 2007). Furthermore, attempts to genetically modify any given Zscan4 locus by conventional gene targeting have been technically hampered due to the highly identical nucleotide sequences of multiple copies of Zscan4 genes and pseudogenes in the Zscan4 genomic cluster. This has been an obstacle for genetic study of the Zscan4 genes.
In this manuscript, we successfully generated ES cell lines and mouse lines with an Emerald-GFP (Emerald) knock-in allele at the Zscan4c locus by using CRISPR/hSpCas9 (Cong et al. 2013) specifically targeting the Zscan4c genomic locus. The established knock-in ES cell lines and mouse lines allowed us to dissect the bona fide expression pattern of Zscan4c and actions of the locus to external stimuli in the context of the endogenous locus in ES cells and two-cell stage embryos. Moreover, combined with mass spectrometry, the knock-in ES cells facilitated analysis of the endogenous Zscan4 protein and its associated factors. Thus, genetically engineered knock-in ES cells at a given Zscan4 locus will shed light on further approaches—not only to study the roles of individual Zscan4 members but also to analyze the knockout of Zscan4 gene clusters in a physiological context.
Materials and Methods
Embryonic stem cell culture
TA1 mouse ES cells (F1 hybrid of C57BL/6J × 129S6/SvEvTac) and the derivative cells were used for all experiments unless otherwise specified (Amano et al. 2013). During the establishment of recombinant ES clones, the cells were initially cultured in 2i+LIF condition (Millipore, Bedford, MA) on the MMC-treated MEF feeder cells. For experiments, ES cell lines were maintained on gelatin-coated feeder-free plates in complete ES medium (Zalzman et al. 2010). For experiments using retinoic acid (RA), all-trans-RA was added at a final concentration of 1 μM in the complete ES medium. Two independent Silencer select siRNA against Zscan4 (Thermo, Kanagawa Japan: s233511, s233512) and negative control siRNA (Thermo: AM4611) were used to prepare Zscan4-depleted and control mESC extracts.
Generation of Zscan4c-Emerald knock-in embryonic stem cells and genotyping
The targeting vector was designed to replace Exon 2 and the following intron of the Zscan4c genomic locus with Emerald-polyA followed by Neo cassette. The targeting arms of 3.56- and 2.66-kb fragments, 5′ and 3′, to the Zscan4c gene, respectively, were generated by PCR from C57BL/6 genomic DNA and directionally cloned in pKOII plasmid, flanking a pGK-Neo-polyA, a loxP site, and a DT-A cassette. The homologous recombinant cells were isolated using TA1 ES cells (F1 hybrid of C57BL/6J × 129S6/SvEvTac) after transfection of the targeting vector together with CRISPR/Cas9 pX330-U6-Chimeric_BB-CBh-hSpCas9 vectors (Addgene, Cambridge, MA) encoding specific guide RNAs 5′-GCCUGUGAUCUGUGGAAGUG-3′ and 5′-CCCACACUUCCACAGAUCAC-3′, which direct the intron between Exon 2 and Exon 3 of the Zscan4c genomic locus. The G418-resistant ES clones were screened for homologous recombination in the Zscan4c locus by PCR using primers, Zscan4c (Z4c)-7206F 5′-AGAATGATATGTGTAGCCATATTAC-3′ and Z4c-Em10892R 5′-AACAGCTCCTCGCCCTTGCTCACCATG-3′ for 5′-arm (3687 bp) and Z4c-Neo-13534F 5′-GCTATCAGGACATAGCGTTGGCTAC-3′ and Z4c-mut16545R 5′-AACAAGCCAAAATTAAAAGGGTTAG-3′ for 3′-arm (3422 bp). Since the Zscan4 genomic cluster consists of multiple copies of Zscan4 genes and pseudogenes with high nucleotide sequence identity, correct targeting at Zscan4c genomic locus was verified by direct sequencing of the PCR products of 5′-arm and 3′-arm regions of the Zscan4c locus for the presence of the specific single-nucleotide differences among Zscan4 loci and fluorescence in situ hybridization (FISH) using a probe encompassing Emerald and Neo genes.
Generation of Zscan4c-Emerald knock-in mice
The chimeric mice were generated by blastocyst injection (host ICR) of recombinant Zscan4c-Emerald-Neo + knock-in ES cells. The male chimeric mice were crossed with C57BL/6J females to generate the F1 offspring. To confirm germline transmission of the Zscan4c-Emerald-Neo+ knock-in allele, genotyping was performed by PCR using primers Z4c-Neo-13534F 5′-GCTATCAGGACATAGCGTTGGCTAC-3′ and Z4c-13786Rmut 5′-CTTGTCCTTGCATTGAAACATGGAC-3′ for 3′-arm (597 bp) and Z4c-10503Fwt 5′-GATACAGGACTTTCTGGTTATTGAG-3′ and Z4c-Em10892R 5′-AACAGCTCCTCGCCCTTGCTCACCATG-3′ for 5′-arm (363 bp). The knock-in mice were congenic with the C57BL/6J background. Whenever possible, each knock-in animal was compared to littermates or age-matched non-littermates from the same colony, unless otherwise described. A transgenic mouse expressing Flippase, D2-Tg(CAG-FLP)18lmeg (from Dr. Masatake Araki, Kumamoto University, CARD), was crossed with a Zscan4c-Emerald-Neo (Neo allele) knock-in mouse to delete pGK-Neo-polyA cassette franked by Frt sequences, producing Zscan4c-Emerald NeoΔ knock-in allele (NeoΔ allele). For genotyping of Zscan4c-Emerald NeoΔ knock-in allele, PCR was performed using primers Z4c-11852F 5′-GGCGGCAGGCCCTGCCATAGCCATG-3′ and Z4c-13786Rmut 5′-CTTGTCCTTGCATTGAAACATGGAC-3′, producing a 261-bp band. Due to the highly identical nucleotide sequences of Zscan4 multigene loci, wild-type Zscan4c locus cannot be distinguished from other Zscan4 loci by PCR genotyping. Thus, to identify homozygous Zscan4c-Emerald knock-in siblings, a heterozygous Zscan4c-Emerald-Neo knock-in mouse (Neo/+) was crossed with a heterozygous Zscan4c-Emerald-NeoΔ knock-in mouse (NeoΔ/+). A homozygous Zscan4c-Emerald knock-in mouse (Neo/NeoΔ) was identified by genomic PCR for the presence of both NeoΔ and Neo allele. Animal experiments were approved by the Institutional Animal Care and Use Committee (approval no. 12702–0).
Embryo collection and culture
Females were injected with 5 U of PMSG (Merck Millipore, Tokyo Japan) and 48 h later with 5 U of hCG (Calbiochem). Embryos were flushed out of the mouse oviducts in pxotassium simplex optimization medium (KSOM) medium 38 h after hCG injection. Embryos were cultured in KSOM medium.
Fluorescence in situ hybridization
ES cells grown on a cover glass were fixed in 4% paraformaldehyde for 5 min; washed with PBS; and subjected to sequential dehydration through 70, 80, 90, and 100% ethanol, denatured in 50% formamide and 2× saline-sodium citrate (SSC) at 72°C for 10 min. Hybridization was conducted with a FITC-labeled probe that detects Emerald GFP and Neo derived from the knock-in vector, in buffer containing 50% formamide, 2× SSC, 20% dextran sulfate, and 0.1 μg/μl of mouse Cot-1 DNA at 37°C for 16 h. The cover glasses were washed sequentially at room temperature in 2× SSC for 1 min, 0.4× SSC/0.3% Tween 20 solution for 2 min, and 2× SSC at room temperature for 1 min. The FITC-labeled probe was generated by nick translation (Hokkaido chromosome science) using equimolar mixture of two templates, Emerald GFP and Neo. DNA fragments of Emerald GFP and Neo were generated by PCR, using primers Z4c-Emerald (Em)-FISH-F 5′-atggtgagcaagggcgaggagctgttc-3′ and Z4c-Em-FISH-R2 5′-gctatggcagggcctgccgccccgacg-3′ for Emerald GFP (1006 bp) and Neo-1F 5′-ATGGGATCGGCCATTGAACAAGATG-3′ and Neo-801R 5′-GAAGAACTCGTCAAGAAGGCGATAG-3′ for Neo (801 bp). Nuclei were counter stained with 4′,6-diamidino-2-phenylindole (DAPI).
In situ hybridization followed by immunostaining
Antisense Zscan4c RNA was synthesized by in vitro transcription with T3 RNA polymerase, using DIG-UTP (Roche, Tokyo Japan) and a mouse Zscan4c complementary DNA (cDNA) template cloned in pBluescript KS+. DIG-labeled antisense Zscan4c RNA probe was fragmented by alkaline (3: 2 mixture of 0.2 M Na2CO3/0.2 M NaHCO3) into 300–500 base at 60°C for 15 min. The ES cells grown on a cover glass were fixed in 4% paraformaldehyde for 5 min, washed with PBS and 0.1% Triton X-100/PBS, and subjected to acetylation by acetic anhydride in 0.1 M triethanolamine (pH 8.0). Hybridization was conducted in 50% formamide, 2× SSC, 1 mg/ml tRNA, 1 mg/ml salmon sperm DNA, 1 mg/ml bovine serum albumin (BSA), and 10% dextran sulfate with DIG-labeled antisense Zscan4c RNA probe at 37°C for 12 h. After washing with NTE buffer (0.5 M NaCl, 10 mM Tris-Cl [pH 8.0], 1 mM EDTA), the unhybridized RNA probe was removed by 20 μg/ml RNase A in NTE buffer. After washing with 0.1× SSC and blocking with 1% digoxigenin (DIG) detection buffer (Roche), immunodetection was done by mouse anti-DIG and rabbit anti-mZscan4 followed by fluorescent second antibodies.
Production of antibodies
Polyclonal antibodies against mouse Zscan4 (aa 1–506) were produced by inserting cDNA fragment in-frame with pET19b (Novagen, Gibbstown, NJ) in Escherichia coli strain BL21-CodonPlus # (DE3). His-tagged recombinant proteins were solubilized in a denaturing buffer (6 M HCl-guanidine, 20 mM Tris-HCl [pH 7.5]) from the inclusion body and purified by Ni-NTA (Qiagen, Hilden, Germany) under denaturing conditions. After dialyzing against PBS, the purified protein was used to immunize rabbits. The antibodies were affinity-purified from the immunized crude serum with immobilized antigen on CNBr-activated Sepharose (GE Healthcare, Piscataway, NJ).
Preparation of chromatin extracts
To prepare chromatin extracts, Emerald-positive mouse ES cells were enriched by fluorescence-activated cell sorting (FACS) sorting and suspended in the extraction buffer (20 mM Tris-HCl [pH 7.5], 100 mM KCl, 0.4 mM EDTA, 0.1% Triton X-100, 10% glycerol, 1 mM β-mercaptoethanol) supplemented with Complete Protease Inhibitor (Roche). After Dounce homogenization, the soluble cytoplasmic/chromatin-unbound fraction was separated after centrifugation at 100,000g for 30 min. The insoluble pellet was washed two times with buffer (10 mM Tris-HCl [pH 7.5], 1 mM CaCl2, 1.5 mM MgCl2, 10% glycerol) and digested with micrococcus nuclease (0.008 units/ml) at 4°C for 60 min. The solubilized fraction was removed after centrifugation for 15 min at 4°C. The chromatin fraction was extracted from the insoluble pellet by high-salt extraction buffer (20 mM HEPES-KOH [pH 7.0], 400 mM KCl, 5 mM MgCl2, 0.1% Tween 20, 10% glycerol, 1 mM β-mercaptoethanol) supplemented with Complete Protease Inhibitor. The solubilized chromatin fraction was collected after centrifugation at 100,000g for 30 min at 4°C.
Immunoprecipitation
For immunoprecipitation of endogenous mouse Zscan4 from chromatin fraction, 5 μg of affinity-purified rabbit anti-mZscan4 and control rabbit IgG antibodies was cross-linked to 50 μl of protein A-Dynabeads by DMP (Sigma). The antibody-cross-linked beads were added to the solubilized chromatin extracts, and the extracts were incubated for 2 h at 4°C with rotation. The beads were washed with high-salt extraction buffer. The bead-bound proteins were eluted with 40 μl of elution buffer (100 mM glycine-HCl [pH 2.5], 150 mM NaCl) and then neutralized with 4 μl of 1 M Tris-HCl [pH 8.0].
Mass spectrometry
For mass spectrometry analysis of the endogenous mouse Zscan4 and its associated proteins, the immunoprecipitates were run on the 10% NuPAGE gel (Thermo Fisher Scientific, Waltham, MA) and separated 2 cm from the well. The gel was excised and washed with 25 mM ammonium bicarbonate followed by acetonitrile. The proteins were reduced with 10 mM DTT at 60°C for 30 min followed by alkylation with 50 mM iodoacetamide and subjected to in-gel digestion by trypsin gold (Promega, Madison, WI) at 37°C for 4 h. The gel digests were analyzed by nano-LC-mass spectrometry (MS)/MS with a Waters NanoAcquity HPLC system interfaced to a Thermo Fisher Orbitrap Velos Pro (MS Bioworks, Ann Arbor, MI). Data were searched using Mascot with the following parameters: database: Swissprot Mouse (forward and reverse appended with common contaminants); fixed modification: carbamidomethyl (C); variable modifications: oxidation (M), acetyl (Protein N-term), Pyro-Glu (N-term Q), and deamidation (NQ); mass values: monoisotopic; peptide mass tolerance 10 ppm; fragment mass tolerance 0.6 Da; and max missed cleavages 2. Mascot DAT files were parsed into the Scaffold software for validation, filtering, and creating a non-redundant list per sample. Data were filtered at 1% protein and peptide false discovery rate and requiring at least two unique peptides per protein. Those proteins were determined as most significantly elevated in the Zscan4 immunoprecipitation (IP) sample versus the control IgG sample based on the following criteria: Protein had at least five spectral counts (SpC) in the Zscan4 IP sample. Protein was not detected in the control IgG sample or detected with a fourfold or more increase based on dividing the SpC values.
Fluorescence-activated cell sorting
The fluorescent intensity analysis and sorting of Emerald of Z4c-Em ES cells were performed using a BD FACSAria II. The cells were sorted according to the fluorescent intensity of Emerald and collected into mouse ES cell culture medium at 4°C.
Immunofluorescence staining
ES cells were grown on a cover glass and fixed in 4% PFA for 5 min at room temperature and permeabilized in 0.1% Triton X-100 in PBS for 10 min. The cells were blocked for 10 min in PBS and 3% BSA and incubated at room temperature with the primary antibodies in a blocking solution. After three washes in PBS, the cells were incubated for 1 h at room temperature with Alexa-dye-conjugated secondary antibodies (1:1000; Invitrogen) in a blocking solution. DNA was counterstained with Vectashield mounting medium containing DAPI (Vector Laboratories).
Imaging
Immunostaining images were captured with DeltaVision and processed with DeltaVision SoftWorx software (GE Healthcare). All the images shown were Z-stacked. Green fluorescent protein (GFP) fluorescence and bright field images were captured with an OLYMPUS IX73 fluorescence microscope and processed with the CellSens standard program.
Antibodies
The following antibodies were used: mouse anti-GFP (Santa Cruz, Santa Cruz, CA: sc-9996), rabbit anti-GFP (Abcam, Cambridge, UK: ab6556), mouse anti-DIG (Abcam: ab6212), and rabbit anti-actin (CST no. 4970).
Results
Infrequent activation of the endogenous mouse Zscan4c promoter in the knock-in embryonic stem cells
Our previous examination of the fluorescent reporter expression from the Zscan4c promoter-Emerald transgene showed that the Zscan4c promoter was transiently activated in only 1–5% of MC1 mouse ES cells (pZscan4-Emerald cell) at a given time point (Zalzman et al. 2010; Akiyama et al. 2015). However, a potential concern was yet to be clarified: whether or not the Zscan4c promoter-Emerald transgene reflects the bona fide expression pattern of Zscan4 locus due to random integration in the genome, copy number effect, and missing cis-regulatory elements in the transgene construct. Thus, we generated an ES cell line in which the Emerald fluorescent reporter is placed at the endogenous Zscan4c locus to examine its expression pattern in the context of the endogenous promoter and regulatory element in ES cells and mouse embryos.
Our previous attempts to generate genetically engineered alleles at any given Zscan4 locus by conventional gene targeting methods failed repeatedly, most likely due to the highly identical nucleotide sequences of multiple copies of Zscan4 paralogs and pseudogenes in the mouse Zscan4 genomic cluster (Fig. 1 A). However, by using CRISPR/hSpCas9 combined with chimeric guide RNAs specifically targeting the Zscan4c genomic locus, we successfully established an ES cell line with Emerald knock-in allele at the Zscan4c locus (Z4c-Emerald-knock-in (KI) ES cell), where Exon 2 and the following intron were replaced by Emerald-polyA (Fig. 1 A). FISH showed that one Zscan4c allele was knocked in, whereas the other Zscan4c allele was intact in the Z4c-Emerald-KI ES cell (Fig. 1 B), indicating that the knock-in ES cell line is heterozygous in terms of Zscan4c allele.
Targeted knock-in of Emerald GFP reporter in Zscan4 gene cluster reveals endogenous activity of Zscan4c locus in ES cells. (A) Schematic illustrations of the 7qA1 region of mouse chromosome 7, where multiple Zscan4 genes (Zscan4a∼f) and pseudogenes (Zscan4-ps1∼3) cluster (arrows: direction of transcription from each Zscan4 gene or pseudogene). The wild-type Zscan4c allele, Zscan4c-Emerald knock-in (Z4c-Emerald-KI) allele, and targeting vector are shown. Targeted replacement of Exon 2 and the following intron of the Zscan4c locus with Emerald GFP results in Z4c-Emerald-KI allele, in which Emerald GFP is under an endogenous Zscan4c promoter. Blue boxes represent exons. Black boxes represent Frt sites. Blue triangles represent lox P sites. Arrowheads indicate the targeting sites of guide RNAs for an hSpCas9-mediated DNA double-strand break. (B) FISH using an FITC-labeled probe that specifically detects the portion of Emerald GFP-Neo in the knock-in allele showed a single dot or a pair of single dots (two juxtaposed signals derived from sister chromatids) before or after the S phase, respectively, indicating that one allele of Zscan4c was replaced by the knock-in vector in the Z4c-Emerald-KI ES clone. Scale bar, 1 μm. (C) Emerald+ cells were infrequently observed in the Z4c-Emerald-KI ES colonies. Left: bright-field image; right: GFP fluorescent image. Scale bars, 50 μm. (D) Fluorescence-activated cell sorting (FACS) analyses of Emerald+ (EM+) ES population in the parental TA1 ES cells (left, negative control), the Z4c-Emerald-KI ES cell clone no. 27 (middle), and MC1 pZ4c-Emerald-Tg ES cells (right) in the conventional (FBS+ LIF) ES culture condition. Note that the overall scatter plot in MC1 pZ4c-Emerald-Tg ES cells shifts toward the right, indicating that the overall Emerald intensity is higher compared to that in Z4c-Emerald-KI ES cells. (E) Zscan4c-Emerald GFP knock-in ES cells were immunostained as indicated using rabbit anti-Zscan4. Arrows: GFP+/Zscan4+ cell; arrowheads: GFP−/Zscan4+ cell; asterisks GFP weakly+/Zscan4 weakly+ cell. Scale bar, 10 μm. Zscan4/GFP-immunostained ES cells were quantified, showing that 1.03% of the total ES cells were Zscan4+ (left graph) and 35.3% of Zscan4-immunopositive cells were GFP+/Zscan4+ (right graph). None of the GFP+/Zscan4-cells was observed. (F) ES cells were subjected to in situ hybridization of bulk Zscan4 mRNA (ISH, red) and immunostaining by rabbit anti-Zscan4 antibody (IF, green), demonstrating that anti-Zscan4 immunopositivity correlates with the expression of bulk Zscan4 mRNA. Note that bulk Zscan4 mRNA is detected over cytosol, while the Zscan4 protein is immunostained in nuclei. Scale bar, 10 μm.
Emerald-positive (Emerald+) cells were infrequently observed in the Z4c-Emerald-KI ES colonies (Fig. 1 C), indicating that the endogenous Zscan4c allele was activated in a limited population. FACS analysis indicated that approximately 0.95% of the Z4c-Emerald-KI ES cells were Emerald+ in the feeder-free conventional mouse ES culture condition, whereas parental cells rarely showed Emerald fluorescence (Fig. 1 D), essentially recapitulating our previous observation that a small subpopulation was Emerald+ in MC1 Z4c-Emerald-transgene (Tg) ES cells (Zalzman et al. 2010). However, we noted that the frequency and fluorescence intensity of Emerald+ cells in the Z4c-Emerald-KI ES cells were less than those observed in MC1 Z4c-Emerald-Tg ES cells (∼2.6%) in the same culture condition (Fig. 1 D). The apparent difference between Z4c-Emerald-KI ES cells and Z4c-Emerald-Tg ES cells could be accounted for partly by the genetic background of mouse ES cell lines: Z4c-Emerald-KI ES cells (F1 hybrid of C57BL/6 and 129) and MC1 Z4c-Emerald-Tg ES cells (129 line), as we have shown previously (Sharova et al. 2007; Amano et al. 2013).
Crucially, coimmunostaining of Z4c-Emerald-KI ES cells by anti-Emerald and anti-Zscan4 antibodies revealed that only a fraction (∼35%) of Zscan4-immunopositive cells showed Emerald immunopositivity (Fig. 1 E), thus indicating that counting Emerald+ cells underestimates the actual total number of Zscan4-positive (Zscan4+) cells. Although we have not assessed precisely the protein turnover of the Emerald GFP in the Z4c-Emerald-KI ES cells, Emerald+ embryonic stem cells (ESCs) always show Zscan4 immunopositivity in the Z4c-Emerald-KI ES cells, indicating that Emerald can monitor the activation of at least endogenous Zscan4c locus in real time. Thus, although anti-Emerald immunofluorescence intensity was correlated with that of anti-Zscan4, it should be noted that Emerald was not expressed in all the Zscan4+ ES cells. This observation seems to be paradoxical, given the evidence that our previous RNA-sequencing analyses showed that all of the Zscan4 paralog genes were expressed in FACS-sorted Emerald+ ES cells (Akiyama et al. 2015). Nevertheless, since our newly generated anti-Zscan4 polyclonal antibody cross-reacts with all of the mouse Zscan4 protein members (at least Zscan4c, Zscan4d, and Zscan4f—see also Fig. 4) and Zscan4 immunopositivity correlates with bulk expression of Zscan4 paralog mRNA (Fig. 1 F), the aforementioned observation rather suggests that the Zscan4c-Emerald knock-in locus is not always activated in a given Zscan4-immunopositive cell. Thus, it is possible that not all of the Zscan4 loci together but a subset of them in the Zscan4 multigene cluster shows transient transcriptional firing at a given time window of a Z4 event in mouse ES cells.
Endogenous mouse Zscan4 genes redundantly function during mouse two-cell stage preimplantation development
Originally, Zscan4 was identified as a gene specifically expressed in the mouse two-cell stage embryo (Falco et al. 2007). Previous RT-PCR suggested that Zscan4d was predominantly expressed among the Zscan4 genes in the two-cell stage embryo, but Zscan4c was also expressed (Falco et al. 2007). To confirm this notion, we generated a knock-in mouse line with a Z4c-Emerald-KI allele from the recombinant ES clone to examine the expression pattern of the endogenous Zscan4c locus in preimplantation embryos. The two-cell stage embryos were collected from heterozygous Z4c-Emerald-KI mice crossed with wild type, half of which should be theoretically heterozygous for Z4c-Emerald-KI. This allowed for a parallel comparison of fluorescence between wild-type and heterozygous Z4c-Emerald-KI embryos in the exact same condition. Although Z4c-Emerald-KI two-cell stage embryos showed less intense fluorescence compared to Emerald+ ES cells, probably due to diffusion over the larger size of the cell, 36% of two-cell embryos showed Emerald fluorescence at 51 h post hCG administration (Fig. 2 A), which is roughly the expected ratio for heterozygous Z4c-Emerald-KI embryos. The number of Emerald+ two-cell embryos increased at 51 h post hCG compared to that at 46 h, suggesting that Z4c-Emerald-KI allele is fully activated in the late rather than the early two-cell stage, consistent with our previous observation by in situ hybridization (Falco et al. 2007). This observation was further confirmed by immunostaining of late two-cell embryos with the anti-Emerald antibody, showing that the Z4c-Emerald-KI late two-cell embryo, but not the associated polar bodies, is immunopositive for Emerald (Fig. 2 B). Thus, the endogenous mouse Zscan4c allele is activated during the embryonic two-cell stage. These results emphasize that the Z4c-Emerald-KI allele recapitulates the stage-specific expression pattern of the endogenous Zscan4c gene in the mouse preimplantation embryo.
The Zscan4c-Emerald GFP reporter knock-in mouse revealed endogenous activities of Zscan4 loci in the mouse preimplantation embryo. (A) GFP fluorescent images of two-cell stage embryos collected from a heterozygous Z4c-Emerald-KI female mouse crossed with a wild-type male at 46 h (upper left) and 51 h (upper right) post hCG. The white arrowhead indicates Emerald+ two-cell embryo. The number of Emerald+ two-cell embryos is summarized in the table (lower). Scale bar, 200 μm. (B) The late two-cell stage embryos at post 51 h hCG were immunostained with anti-GFP. Z4c-Emerald KI two-cell embryo showed GFP staining, while polar bodies or wild-type two-cell embryos did not. Scale bar, 50 μm. (C) Schematic illustrations of the wild-type Zscan4c allele (+), Zscan4c-Emerald GFP-Neo knock-in allele (Neo), and Zscan4c-Emerald GFP-NeoΔ knock-in allele (NeoΔ) are shown (upper). The NeoΔ allele knock-in mouse line was generated by crossing a Neo knock-in mouse with a transgenic mouse expressing Flippase to delete pGK-Neo-polyA franked by Frt sequences. Arrows: PCR primes. Due to the highly identical nucleotide nature of Zscan4 multigene loci, wild-type Zscan4c locus cannot be distinguished from other Zscan4 loci by PCR genotyping. To facilitate identification of homozygous Zscan4c-Emerald knock-in siblings, Neo/+ mouse was crossed with NeoΔ/+, producing homozygous Neo/NeoΔ, which was identified by PCR (lower left). Homozygous Neo/NeoΔ siblings, in which functional Zscan4c allele is disrupted, are viable (lower right).
Importantly, we noted that mice with a homozygous Zscan4c-Emerald-KI allele, which is supposed to be a knockout of functional Zscan4c genes, were still viable without any obvious defect (Fig. 2 C). Moreover, there is no overt phenotypic difference in the reproduction and development between heterozygous and homozygous knock-in mice (Fig. 2 C), suggesting that loss of functional Zscan4c genes was compensated by other Zscan4 loci—most likely Zscan4d—during embryonic development. Thus, it is suggested that not only Zscan4c but also other Zscan4 genes act redundantly for preimplantation embryonic development.
The endogenous mouse Zscan4c promoter is activated by retinoic acid in the knock-in embryonic stem cells
It has been demonstrated that expression of Zscan4 is positively regulated by telomere shortening (Nakai-Futatsugi and Niwa 2016), DNA damage, and PI3 kinase-mediated external signals (Storm et al. 2014) and is negatively regulated by Tet (Lu et al. 2014), Rif1 (Dan et al. 2014), and Nr01b (Fujii et al. 2015). However, the molecular details of how an infrequent burst of endogenous Zscan4 expression is triggered remain largely unknown.
Our previous study demonstrated that the burst of transcription at Z4 events coincides with transient histone hyperacetylation (Akiyama et al. 2015). Thus, we examined whether an HDAC inhibitor triggers activation of the Z4c-Emerald knock-in locus. FACS analysis indicated that Emerald+ ES cells were slightly increased (Fig. 3 A, B compared with Fig. 1 D) in the presence of the HDAC inhibitor, valproic acid (VPA), suggesting that histone acetylation slightly promotes activation of the Z4c-Emerald-KI locus.
The Zscan4c-Emerald GFP reporter knock-in locus is activated by retinoic acid in ES cells. (A) GFP fluorescence images of the Z4c-Emerald-KI ES cells in the presence of VPA (0.5 μM) or RA (1 μM, 10 μM) and of the MC1 ES cells with Z4c-GFP-IRES-Puro transgene in the presence of RA (10 μM) and puromycin. Scale bar, 200 μm. (B) FACS analyses of the GFP+ population in the Z4c-Emerald-KI ES cells and MC1 Z4c-GFP-IRES-Puro Tg ES cells as indicated in (A). Note that the overall scatter plot in Z4c-GFP-IRES-Puro Tg ES cells, which acquire puromycin resistance only when GFP is expressed, shifts toward the right, indicating that the overall GFP intensity of the survived cells selected against puromycin is higher compared to that in Z4c-Emerald-KI ES cells.
Given that retinoic acid (RA) activates Zscan4 expression (Sharova et al. 2007) (Sharova et al. 2016), we examined the expression of Emerald from Z4c-Emerald-KI locus after RA treatment. Remarkably, FACS analysis indicated that approximately 16–17% of the Z4c-Emerald-KI ES cells were Emerald+ in the presence of RA for 48 h (Fig. 3 A, B), suggesting that the endogenous Zscan4c locus is directly or indirectly activated in response to RA. To exclude a possibility of auto-fluorescence derived from dead or dying cells, we assessed the RA effect on MC1 ES cells with the Zscan4c promoter-EGFP-IRES-puro transgene (MC1 Z4c-EGFP-IRES-Puro Tg ES cell), which acquire puromycin resistance upon EGFP expression (Fig. 3 A, B). Because the Zscan4c-activated cells survived against puromycin selection and showed EGFP fluorescence with high intensity, we can exclude a possibility that dying or dead cells with auto-fluorescence were detected after RA treatment. Although physiological relevance of RA signaling for activation of the endogenous Zscan4 locus requires further investigation, the endogenous Zscan4 locus might have a cis-regulatory element that responds to RA or relevant stimuli. Thus, the Zscan4 locus acts as a sensor to cell-extrinsic and cell-intrinsic stimuli in ES cells.
Expression of endogenous Zscan4 proteins and their associated factors in embryonic stem cells
Our previous RNA-sequencing analyses showed that all the Zscan4 genes were activated during the Z4 event (Akiyama et al. 2015). Due to limited available material, however, it remained elusive which members of the endogenous Zscan4 proteins are expressed in ES cells. Moreover, Zscan4 proteins show high amino acid sequence identity, which hampers the technical ability to identify an individual Zscan4 protein member by anti-Zscan4 antibody, which was previously raised against the most C-terminal peptide (14aa) epitope of Zscan4c (Zalzman et al. 2010). Nevertheless, the evidence that RA enhances Emerald reporter expression from the knock-in allele prompted us to collect Emerald+ ES cells by FACS sorting and examine the endogenous Zscan4 protein. For this analysis, we generated new polyclonal antibodies against the entire full length (506 aa) of the Zscan4 protein since the previously raised anti-Zscan4 antibody (Zalzman et al. 2010) may have missed detection of the truncated types of Zscan4a, Zscan4b, and Zscan4e proteins. Our biochemical fractionation followed by immunoprecipitation demonstrated that the endogenous Zscan4 protein was mostly separated into the chromatin-bound fraction extracted from Emerald+ ES cells (Fig. 4 A), suggesting that the endogenous Zscan4 protein is tightly associated with chromatin rather than free from it.
Mass spectrometry analysis revealed the expression of endogenous Zscan4 proteins and their associated factors in Z4c-Emerald knock-in ES cells. (A) Emerald+ mouse ES cells after RA treatment for 48 h were separated into chromatin bound and unbound fractions. Immunoprecipitation (IP) was done by control IgG and rabbit anti-Zscan4 antibody from chromatin-bound and chromatin-unbound fractions. Western blotting showed that endogenous Zscan4 protein was exclusively extracted from the chromatin-bound fraction. (B) Silver-stained gel showing immunoprecipitates (IP) by control IgG and rabbit anti-Zscan4 antibody from the chromatin fraction of Emerald+ ES cells, enriched by FACS sorting after RA treatment. M: molecular weight marker. Arrowhead: Zscan4. (C) LC-MS/MS analysis of immunoprecipitated endogenous Zscan4 showing unique MS spectrums specifically assigned to peptide sequences derived from Zscan4c, Zscan4d, or Zscan4f. (D) Western blotting of immunoprecipitated endogenous Zscan4 probed with rabbit anti-Zscan4 antibody. Arrowhead: Zscan4. Asterisk: IgG heavy chain. (E) Whole-cell extracts of RA-treated Z4c-Emerald KI ES cells after administration of Zscan4 or negative control siRNA were probed with antibodies as indicated. Note that specificity of the immunoreactive band detected by rabbit anti-Zscan4 antibody was confirmed by siRNA against Zscan4 genes. (F) List of proteins identified in the Zscan4 immunoprecipitates by LC-MS/MS analysis. Spectral counts (SpC) of the identified peptide are indicated. Highlighted in red are the Zscan4-associated factors identified in the previous MS analysis of Zscan4c-FLAG immunoprecipitates after overexpression (Akiyama et al. 2015).
Intriguingly, MS analysis of the immunoprecipitated endogenous Zscan4 identified unique MS spectrums, which were specifically assigned to trypsin-digested peptide sequences derived from Zscan4c, Zscan4d, or Zscan4f (Fig. 4 B, C). Notably, the identified MS spectrums showed more Zscan4c peptides than other Zscan4 peptides (Fig. 4 C), suggesting that the Zscan4c protein could be dominantly expressed among the Zscan4 proteins in the Emerald+ ES cell population, in agreement with predominant expression of Zscan4c mRNA (Falco et al. 2007). By contrast, this analysis identified none of the peptides assigned to truncated type members (Zscan4a, Zscan4b, or Zscan4e), despite redundant coverage of peptides from Zscan4c, Zscan4d, or Zscan4f. Consistent with this observation, immunoprecipitation followed by Western blotting of Zscan4 showed that only a single band, which corresponds to larger forms (Zscan4c, Zscan4d, or Zscan4f) but not to putative truncated forms (Zscan4a, Zscan4b, or Zscan4e), was detectable (Fig. 4 D). Moreover, siRNAs against Zscan4 genes confirmed that this immunoreactive band is specific to Zscan4 (Fig. 4 E). Thus, these results suggest that a subset of different Zscan4 members is indeed expressed at a protein level in Emerald+ ES cells, while expression of the putative truncated form of Zscan4 is less than detectable level.
A previous overexpression study using a doxycycline-inducible transgene of FLAG-Zscan4c showed that Zscan4c is associated with transcriptional repressors and activators (Storm et al. 2014; Akiyama et al. 2015). However, factors associated with endogenous Zscan4 are yet to be analyzed despite physiological relevance. Our MS analysis of the immunoprecipitated endogenous Zscan4 identified potential Zscan4-associated factors (Fig. 4 F). Consistent with our previous report using the exogenous FLAG-Zscan4c (Storm et al. 2014; Akiyama et al. 2015), the data indicate that transcriptional repressors KDM1A/LSD1, KAP1/TIF1β, and HDAC1 are associated with endogenous Zscan4, supporting our previous observation that Zscan4 acts for transcriptional repression at heterochromatic regions (Akiyama et al. 2015).
Discussion
The repetitive nature of the multiple Zscan4 genes in mouse and their strictly limited expression patterns have hampered genetic analysis of a Zscan4 locus in physiological conditions. Here, the CRISPR/Cas9 technique, specifically directed at a Zscan4c locus, allowed us to scrutinize bona fide expression patterns and mode of action to external stimuli of endogenous Zscan4c gene in ES cells and two-cell stage embryos. Because we employed conventional CRISPR/Cas9 system in this study, we cannot exclude a possibility of off-target cuts and indels in the established ESC clones. However, it is unlikely that this concern affects our analyses of Zscan4 locus, because (i) different clones of the knock-in ESC essentially recapitulated the same character in terms of Emerald expression and (ii) the established knock-in mouse lines were viable.
Intriguingly, our study uncovered that the Zscan4c-Emerald knock-in locus is not always activated in a given Zscan4+ ES cell (Fig. 1 F), suggesting that a subset of Zscan4 loci in the Zscan4 multigene cluster is expressed at a given time of the Z4 event in mouse ES cells. It was previously shown that expression of Zscan4 is activated by DNA damage, PI3 kinase-mediated signal (Storm et al. 2014), and RAs (Sharova et al. 2007; Sharova et al. 2016) and suppressed by DNA cytosine demethylation status (Lu et al. 2014), telomeric factor Rif1 (Dan et al. 2014), and nuclear receptor Nr01b (Fujii et al. 2015). Similarly, we demonstrated that endogenous Zscan4 expression is triggered in response to extrinsic stimuli in ES cells (Fig. 3). Therefore, the implication for those observations is that the Zscan4 locus is imposed under several layers of regulations that respond to multiple intrinsic and extrinsic stimuli in ES cells and probably during ZGA in two-cell stage embryos. Thus, the molecular details regarding how the burst of endogenous Zscan4 expression is acquired in ES cells and embryos await further investigation.
Supporting previous overexpression studies (Akiyama et al. 2015), we have demonstrated that the endogenous Zscan4 protein is bound to chromatin in association with KDM1A/LSD1, KAP1/TIF1β, and HDAC1 (Fig. 4), as has been implicated in the rapid derepression and rerepression of heterochromatin regions (Akiyama et al. 2015). It is worth noting that robust phosphorylation of KAP1/TIF1β by ATM is a critical determinant for 53BP1-mediated DNA damage repair in the heterochromatic region, which is otherwise inhibitory to repair (Noon et al. 2010). Given that the Zscan4+ cell is accompanied by arrest or accumulation at the G2 phase of the cell cycle in ES cells (Nakai-Futatsugi and Niwa 2016) (Storm et al. 2014) and late two-cell stage embryos (Fig. 2) (Falco et al. 2007), it is possible that Zscan4 plays pivotal roles not only in transcriptional regulation but also in a checkpoint response to heterochromatic regions for stable genome integrity.
Taken together, our genetically engineered knock-in ES cells provided insight into modes of action at a Zscan4 locus. Combining with CRISPR/Cas9-mediated targeting at a different Zscan4 gene, e.g., Zscan4d, in this locus will allow us not only to study the biological roles of individual Zscan4 members but also to analyze the knockout phenotype of the Zscan4 gene cluster in ES cells and preimplantation embryos.
Conclusion
We newly generated knock-in mouse ES cell lines and mouse lines, in which the expression of endogenous Zscan4c, one of the Zscan4 genes, can be specifically monitored with a green fluorescent protein. Our genetic engineering at the endogenous Zscan4c gene allowed the first analysis for the expression and function of each gene copy of Zscan4 locus in a physiological context, providing previously unforeseen phenomena by Zscan4-transgene studies. Our data showed that only ∼30% of Zscan4-immunopositive ES cells were Emerald positive, suggesting that even when a given Zscan4 locus is active, not all endogenous Zscan4 genes are expressed synchronously. Our MS of protein complexes associated with endogenous Zscan4 proteins showed that Zscan4 acts as a transcriptional repressor in a physiological context.
References
Akiyama T, Xin L, Oda M, Sharov AA, Amano M, Piao Y, Cadet JS, Dudekula DB, Qian Y, Wang W, et al. (2015) Transient bursts of Zscan4 expression are accompanied by the rapid derepression of heterochromatin in mouse embryonic stem cells. DNA Res
Amano T, Hirata T, Falco G, Monti M, Sharova LV, Amano M, Sheer S, Hoang HG, Piao Y, Stagg CA, et al. (2013) Zscan4 restores the developmental potency of embryonic stem cells. Nat Commun 4:1966
Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823
Dan J, Liu Y, Liu N, Chiourea M, Okuka M, Wu T, Ye X, Mou C, Wang L, Wang L, et al. (2014) Rif1 maintains telomere length homeostasis of ESCs by mediating heterochromatin silencing. Dev Cell 29:7–19
Falco G, Lee SL, Stanghellini I, Bassey UC, Hamatani T, Ko MS (2007) Zscan4: a novel gene expressed exclusively in late 2-cell embryos and embryonic stem cells. Dev Biol 307:539–550
Fujii S, Nishikawa-Torikai S, Futatsugi Y, Toyooka Y, Yamane M, Ohtsuka S, Niwa H (2015) Nr0b1 is a negative regulator of Zscan4c in mouse embryonic stem cells. Sci Rep 5:9146
Hirata T, Amano T, Nakatake Y, Amano M, Piao Y, Hoang HG, Ko MS (2012) Zscan4 transiently reactivates early embryonic genes during the generation of induced pluripotent stem cells. Sci Rep 2:208
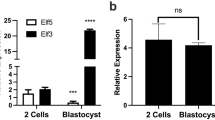
Hung SS, Wong RC, Sharov AA, Nakatake Y, Yu H, Ko MS (2013) Repression of global protein synthesis by Eif1a-like genes that are expressed specifically in the two-cell embryos and the transient Zscan4-positive state of embryonic stem cells. DNA Res 20:391–402
Jiang J, Lv W, Ye X, Wang L, Zhang M, Yang H, Okuka M, Zhou C, Zhang X, Liu L, et al. (2013) Zscan4 promotes genomic stability during reprogramming and dramatically improves the quality of iPS cells as demonstrated by tetraploid complementation. Cell Res 23:92–106
Lu F, Liu Y, Jiang L, Yamaguchi S, Zhang Y (2014) Role of Tet proteins in enhancer activity and telomere elongation. Genes Dev 28:2103–2119
Nakai-Futatsugi Y, Niwa H (2016) Zscan4 is activated after telomere shortening in mouse embryonic stem cells. Stem Cell Rep 6:483–495
Noon AT, Shibata A, Rief N, Lobrich M, Stewart GS, Jeggo PA, Goodarzi AA (2010) 53BP1-dependent robust localized KAP-1 phosphorylation is essential for heterochromatic DNA double-strand break repair. Nat Cell Biol 12:177–184
Sharova LV, Sharov AA, Piao Y, Shaik N, Sullivan T, Stewart CL, Hogan BL, Ko MS (2007) Global gene expression profiling reveals similarities and differences among mouse pluripotent stem cells of different origins and strains. Dev Biol 307:446–459
Sharova LV, Sharov AA, Piao Y, Stagg CA, Amano T, Qian Y, Dudekula D, Schlessinger D, and Ko MS (2016) Emergence of undifferentiated colonies from mouse embryonic stem cells undergoing differentiation by retinoic acid treatment. In Vitro Cell Dev Biol Anim
Storm MP, Kumpfmueller B, Bone HK, Buchholz M, Sanchez Ripoll Y, Chaudhuri JB, Niwa H, Tosh D, Welham MJ (2014) Zscan4 is regulated by PI3-kinase and DNA-damaging agents and directly interacts with the transcriptional repressors LSD1 and CtBP2 in mouse embryonic stem cells. PLoS One 9:e89821
Zalzman M, Falco G, Sharova LV, Nishiyama A, Thomas M, Lee SL, Stagg CA, Hoang HG, Yang HT, Indig FE, et al. (2010) Zscan4 regulates telomere elongation and genomic stability in ES cells. Nature 464:858–863
Acknowledgments
We thank the Collaborative Research Resources, School of Medicine, Keio University, for technical assistance and all members of the Ko laboratory for their support and discussion. This work was supported in part by a Grant-in-Aid for Scientific Research on Innovative Areas “Mechanisms regulating gamete formation in animals” (KAKENHI no. 16H01257); a Grant-in-Aid for Scientific Research on Innovative Areas “Epigenome dynamics and regulation in germ cells” (KAKENHI no. 16H01221); a Grant-in-Aid for Scientific Research (C) (KAKENHI no. 26440003); a Grant-in-Aid for Challenging Exploratory Research (KAKENHI no. 15606842) from MEXT, Japan; the Takeda Science Foundation and the Uehara Memorial Foundation; the Mochida Memorial Foundation for Medical and Pharmaceutical Research and the Yamada Science Foundation (to K.I.); and a Grant-in-Aid for Challenging Exploratory Research (KAKENHI no. 15606842) from MEXT, Japan (to M.S.H.K.), a Grant-in-Aid Scientific Research (B) (KAKENHI no. 15H04812), and a Grant-in-Aid for challenging Exploratory Research (KAKENHI no. 16K15433) from MEXT, Japan (to S.B.H.K.). Animal experiments were approved by the Institutional Animal Care and Use Committee (approval no. 12702-0).
Author contribution
K.I. designed study, conducted experiments, and wrote the manuscript. Y.N. generated Z4-EGFP-IRES-puro MC1 ES cells. H.K., N.C., M.O., T.A., and S.B.H.K. contributed to data acquisition. M.S.H.K. conceived and designed the project and wrote the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
M.S.H.K. is a cofounder of Elixirgen, LLC, which promotes clinical application of human Zscan4.
Additional information
Editor: Tetsuji Okamoto
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Ishiguro, Ki., Nakatake, Y., Chikazawa-Nohtomi, N. et al. Expression analysis of the endogenous Zscan4 locus and its coding proteins in mouse ES cells and preimplantation embryos. In Vitro Cell.Dev.Biol.-Animal 53, 179–190 (2017). https://doi.org/10.1007/s11626-016-0097-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11626-016-0097-y