Abstract
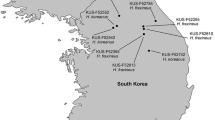
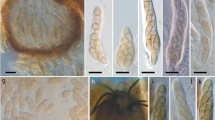
Hymenoscyphus is a large fungal genus containing a single known severe pathogen (H. fraxineus) causing ash dieback in Europe. Here, the closely related species Hymenoscyphus linears sp. nov. is described from Japan. Apothecia of this fungus emerge from linear, black pseudosclerotia on rachises and petioles of Fraxinus platypoda. In culture, the species forms a Chalara anamorph similar to that of H. fraxineus. In addition, a Sporotrichum-like synanamorph is produced. Spores of neither anamorphic forms germinate on malt extract agar and might act solely as spermatia. After prolonged incubation, ascocarps are produced directly on agar plates, indicating the ability to self-fertilize. Phylogenetic investigations using four sequence markers placed the species alongside H. albidus, H. albidoides, and H. fraxineus. Eight fungal strains, co-occurring with H. linearis on petioles of F. platypoda, were isolated and identified by sequencing the ITS region of the rDNA. Seven different species were revealed, showing that there is no other dominant fungus on petioles of F. platypoda. Pathogenicity tests on Fraxinus excelsior and its native host F. platypoda revealed that the fungus is avirulent. The close phylogenetic relationship with H. fraxineus and the avirulence of H. linearis offer an opportunity to study the evolution of pathogenicity of H. fraxineus in further detail in the future.




Similar content being viewed by others
References
Anonymous (2005) Pantone color bridge/coated. Pantone Inc., New Jersey
Baral HO (1996) Hymenoscyphus seminis-alni, a new species of the H. fructigenus-complex. Mycotaxon 60:249–256
Baral HO, Arenal F, Collado J, Galán R, López J, Peláez F, Platas G, Rubio V, Villarreal M (2006) Hynenoscyphus crataegi (Helotiales), a new species from Spain and its phylogenetic position within the genus Hymenoscyphus. Sydowia 58(2):145–162
Baral HO, Galán R, Platas G, Tena R (2013a) Phaeohelotium undulatum comb. nov. and Phaeoh. succineoguttulatum sp. nov., two segregates of the Discinella terrestris aggregate found under Eucalyptus in Spain: taxonomy, molecular biology, ecology and distribution. Mycosystema 32(3):386–428
Baral HO, Marson G, Bogale M, Untereiner WA (2013b) Xerombrophila crystallifera, a new genus and species in the Helotiales. Mycol Progr 12(3):475–488. doi:10.1007/s11557-012-0854-6
Baral HO, Queloz V, Hosoya T (2014) Hymenoscyphus fraxineus, the correct scientific name for the fungus causing ash dieback in europe. IMA Fungus 5(1):79–80. doi:10.5598/imafungus.2014.05.01.09
Blair C, Murphy RW (2011) Recent trends in molecular phylogenetic analysis: Where to next? J Hered 102(1):130–138. doi:10.1093/jhered/esq092
Brasier CM (2000) Plant pathology: The rise of the hybrid fungi. Nature 405(6783):134–135
Brasier CM, Kirk SA (2010) Rapid emergence of hybrids between the two subspecies of Ophiostoma novo-ulmi with a high level of pathogenic fitness. Plant Pathol 59(1):186–199
Carbone I, Kohn LM (1999) A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91(3):553–556. doi:10.2307/3761358
Carmichael JW (1980) Genera of Hyphomycetes. Press, University of Alberta
Drenkhan R, Hanso M (2010) New host species for Chalara fraxinea. New Disease Reports 22:16
Drummond A, Rambaut A (2008) Tracer [Online]. Available at: http://beast.bio.ed.ac.uk/Tracer (accessed 25. October 2013).
Et-Touil A, Brasier CM, Bernier L (1999) Localization of a pathogenicity gene in Ophiostoma novo-ulmi and evidence that it may be introgressed from O. ulmi. Mol Plant-Microbe Interact 12(1):6–15. doi:10.1094/MPMI.1999.12.1.6
Giraud T, Gladieux P, Gavrilets S (2010) Linking emergence of fungal plant diseases and ecological speciation. Trends Ecol Evol 25(7):387–395. doi:10.1016/j.tree.2010.03.006
Gross A, Holdenrieder O (2013) On the longevity of Hymenoscyphus pseudoalbidus in petioles of Fraxinus excelsior. Forest Pathol 43(2):168–170. doi:10.1111/efp.12022
Gross A, Zaffarano PL, Duo A, Grünig CR (2012) Reproductive mode and life cycle of the ash dieback pathogen Hymenoscyphus pseudoalbidus. Fungal Genet Biol 49(12):977–986. doi:10.1016/j.fgb.2012.08.008
Gross A, Holdenrieder O, Pautasso M, Queloz V, Sieber TN (2014a) Hymenoscyphus pseudoalbidus, the causal agent of ash dieback. Mol Plant Pathol 15(1):109–117
Gross A, Hosoya T, Queloz V (2014b) Population structure of the invasive forest pathogen Hymenoscyphus pseudoalbidus. Mol Ecol 23(12):2943–2960. doi:10.1111/mec.12792
Grünig CR, Brunner PC, Duò A, Sieber TN (2007) Suitability of methods for species recognition in the Phialocephala fortinii–Acephala applanata species complex using DNA analysis. Fungal Genet Biol 44(8):773–788
Han J-G, Shin H–D (2008) Hymenoscyphus ginkgonis sp. nov. growing on leaves of Ginkgo biloba. Mycotaxon 103:189–195
Hosoya T, Otani Y, Furuya K (1993) Materials for the fungus flora of Japan (46). T Mycol Soc Jpn 34(4):429–432
Kirisits T, Dämpfle L, Kräutler K (2013) Hymenoscyphus albidus is not associated with an anamorphic stage and displays slower growth than Hymenoscyphus pseudoalbidus on agar media. Forest Pathol. doi:10.1111/efp.12042
Kirk PM, Cannon PF, Minter DW, Stalpers JA (2008) Ainsworth and Bisby's dictionary of the fungi. In, vol 10th edition. CAB International, Wallingford, p 771
Kowalski T, Holdenrieder O (2009) The teleomorph of Chalara fraxinea, the causal agent of ash dieback. Forest Pathol 39(5):304–308
Lanfear R, Calcott B, Ho SYW, Guindon S (2012) PartitionFinder: Combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol 29(6):1695–1701. doi:10.1093/molbev/mss020
Marčiulynienė D, Cleary MR, Shabunin D, Stenlid J, Vasaitis R Detection of Hymenoscyphus pseudoalbidus in Primorye region, far east Russia; COST Action FP1103 Fraxback 4th MC meeting & workshop “Frontiers in ash dieback research”, 4–6th of September 2013, Malmö, Sweden; accessed online 21.01.2014 http://www.fraxback.eu/. In, 2013. p 42
McKinney LV, Nielsen LR, Collinge DB, Thomsen IM, Hansen JK, Kjær ED (2014) The ash dieback crisis: genetic variation in resistance can prove a long term solution. Plant Pathol 63(3):485–499. doi:10.1111/ppa.12196
Olson Å, Stenlid J (2002) Pathogenic fungal species hybrids infecting plants. Microb Infect 4(13):1353–1359
Pautasso M, Aas G, Queloz V, Holdenrieder O (2013) European ash (Fraxinus excelsior) dieback — A conservation biology challenge. Biol Conserv 158:37–49. doi:10.1016/j.biocon.2012.08.026
Plattner A, Kim J-J, Reid J, Hausner G, Lim YW, Yamaoka Y, Breuil C (2009) Resolving taxonomic and phylogenetic incongruence within species Ceratocystiopsis minuta. Mycologia 101(6):878–887. doi:10.3852/08-132
Queloz V, Grünig CR, Berndt R, Kowalski T, Sieber TN, Holdenrieder O (2011) Cryptic speciation in Hymenoscyphus albidus. Forest Pathol 41(2):133–142. doi:10.1111/j.1439–0329.2010.00645.x
Rintoul TL, Eggertson QA, Levesque CA (2012) Multigene phylogenetic analyses to delimit new species in fungal plant pathogens. In: Bolton MD, Thomma B (eds) Plant Fungal Pathogens: Methods and Protocols, vol 835. Methods in Molecular Biology. pp 549–569. doi:10.1007/978–1–61779–501–5_34
Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP (2012) MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61(3):539–542. doi:10.1093/sysbio/sys029
Slippers B, Stenlid J, Wingfield MJ (2005) Emerging pathogens: fungal host jumps following anthropogenic introduction. Trends Ecol Evol 20(8):420–421. doi:10.1016/j.tree.2005.05.002
Sokolski S, Piché Y, Bérubé JA (2004) Lophodermium macci sp. nov., a new species on senesced foliage of five-needle pines. Mycologia 96(6):1261–1267
Stamatakis A (2006) RAxML–VI–HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22(21):2688–2690. doi:10.1093/bioinformatics/btl446
Stukenbrock EH (2013) Evolution, selection and isolation: a genomic view of speciation in fungal plant pathogens. New Phytol 199(4):895–907. doi:10.1111/nph.12374
Taylor J, Swann E (1994) DNA from herbarium specimens. In: Hummel S (ed) Herrmann B. Ancient DNA, Springer New York, pp 166–181. doi:10.1007/978-1-4612-4318-2_11
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA et al (eds) PCR protocols: A guide to methods and applications. Academic Press, Inc., San Diego, USA and London, England, pp 315–322
Wiens JJ, Morrill MC (2011) Missing data in phylogenetic analysis: Reconciling results from simulations and empirical data. Syst Biol. doi:10.1093/sysbio/syr025
Yang ZL (2000) Revision of the Chinese Amanita collections deposited in BPI and CUP. Mycotaxon 75:117–130
Zhang Y-H, Zhuang W-Y (2004) Phylogenetic relationships of some members in the genus Hymenoscyphus (Ascomycetes, Helotiales). Nova Hedwigia 78(3–4):475–484. doi:10.1127/0029-5035/2004/0078-0475
Zhao Y-J, Hosoya T, Baral H-O, Hosaka K, Kakishima M (2012) Hymenoscyphus pseudoalbidus, the correct name for Lambertella albida reported from Japan. Mycotaxon 122:25–41. doi:10.5248/122.25
Zheng H-D, Zhuang W-Y (2013) Hymenoscyphus albidoides sp. nov. and H. pseudoalbidus from China. Mycol Progr 13(3):625–638. doi:10.1007/s11557-013-0945-z
Acknowledgments
We would like to thank N. Kamata for guidance in the Chichibu forest and W. Gams for his help regarding the description of the synanamorph. Further thanks go to O. Holdenrieder, T. Sieber and V. Queloz for interesting discussions and suggestions to M. Berchtold and S. Stroheker for excellent technical assistance and to C. Syrad for manuscript proofreading. We would also like to extend our thanks to the Genetic Diversity Center of ETH Zurich for providing laboratory facilities. This study was supported by a grant from the ETH Zurich (ETH–04 10–1).
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Online Resource 1
Primers used to amplify the sequence markers of the different Hymenoscyphus species. NA indicates unavailable data. (XLSX 10 kb)
Online Resource 2
Maximum likelihood single-gene trees of (a) the ITS rDNA, (b) partial actin, (c) partial calmodulin and (d) partial elongation factor 1-α genes. ML bootstrap percentages are given at the nodes. The scale bar below the trees represents the number of substitutions per site. (GIF 289 kb)
High resolution image
(TIFF 2233 kb)
Rights and permissions
About this article
Cite this article
Gross, A., Hosoya, T., Zhao, YJ. et al. Hymenoscyphus linearis sp. nov: another close relative of the ash dieback pathogen H. fraxineus . Mycol Progress 14, 20 (2015). https://doi.org/10.1007/s11557-015-1041-3
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11557-015-1041-3