Abstract
Purpose
Template-based segmentation techniques have been developed to facilitate the accurate targeting of deep brain structures in patients with movement disorders. Three template-based brain MRI segmentation techniques were compared to determine the best strategy for segmenting the deep brain structures of patients with Parkinson’s disease.
Methods
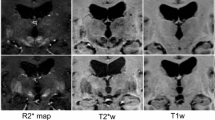
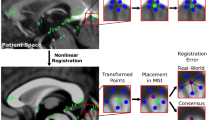
T1-weighted and T2-weighted magnetic resonance (MR) image templates were created by averaging MR images of 57 patients with Parkinson’s disease. Twenty-four deep brain structures were manually segmented on the templates. To validate the template-based segmentation, 14 of the 24 deep brain structures from the templates were manually segmented on 10 MR scans of Parkinson’s patients as a gold standard. We compared the manual segmentations with three methods of automated segmentation: two registration-based approaches, automatic nonlinear image matching and anatomical labeling (ANIMAL) and symmetric image normalization (SyN), and one patch-label fusion technique. The automated labels were then compared with the manual labels using a Dice-kappa metric and center of gravity. A Friedman test was used to compare the Dice-kappa values and paired t tests for the center of gravity.
Results
The Friedman test showed a significant difference between the three methods for both thalami (p < 0.05) and not for the subthalamic nuclei. Registration with ANIMAL was better than with SyN for the left thalamus and was better than the patch-based method for the right thalamus.
Conclusion
Although template-based approaches are the most used techniques to segment basal ganglia by warping onto MR images, we found that the patch-based method provided similar results and was less time-consuming. Patch-based method may be preferable for the subthalamic nucleus segmentation in patients with Parkinson’s disease.
Similar content being viewed by others
References
Benabid AL, Chabardes S, Mitrofanis J, Pollak P (2009) Deep brain stimulation of the subthalamic nucleus for the treatment of Parkinson’s disease. Lancet Neurol 8: 67–81
Koller W, Pahwa R, Lyons K, Wilkinson S (2000) Deep brain stimulation of the Vim nucleus of the thalamus for the treatment of tremor. Neurology 55: S29–S33
Vayssiere N, Hemm S, Zanca M, Picot MC, Bonafe A, Cif L, Frerebeau P, Coubes P (2000) Magnetic resonance imaging stereotactic target localization for deep brain stimulation in dystonic children. J Neurosurg 93: 784–790
Parsons TD, Rogers SA, Braayen AJ, Woods SP, Troster AI (2006) Cognitive sequelae of subthalamic nucleus deep brain stimulation in Parkinson’s disease: a meta-analysis. Lancet Neurol 5: 578–588
York MK, Wilde EA, Simpson R, Jankovic J (2009) Relationship between neuropsychological outcome and DBS surgical trajectory and electrode location. J Neurol Sci 287: 159–171
Bardinet E, Bhattacharjee M, Dormont D, Pidoux B, Malandain G, Schüpbach M, Ayache N, Cornu P, Agid Y, Yelnik J (2009) A three-dimensional histological atlas of the human basal ganglia. II. Atlas deformation strategy and evaluation in deep brain stimulation for Parkinson disease. J Neurosurg 110: 208–219
Chakravarty MM, Bertrand G, Hodge CP, Sadikot AF, Collins DL (2006) The creation of a brain atlas for image guided neurosurgery using serial histological data. Neuroimage 30: 359–376
Lalys F, Haegelen C, Ferre JC, El-Ganaoui O, Jannin P (2010) Construction and assessment of a 3T-MRI brain template. Neuroimage 49: 345–354
Schaltenbrand G, Wahren W (1977) Atlas for stereotaxy of the human brain. Thieme, Stuttgart
Talairach J, Tournoux P (1988) Co-planar stereotaxic atlas of the human brain. Georg Thieme Verlag, Stuttgart
Yelnik J, Bardinet E, Dormont D, Malandain G, Ourselin S, Tandé D, Karachi K, Ayache N, Cornu P, Agid Y (2007) A three-dimensional, histological and deformable atlas of the human basal ganglia. I. Atlas construction based on immunohistochemical and MRI data. Neuroimage 34: 618–638
D’Haese PF, Pallavaram S, Li R, Remple MS, Kao C, Neimat JS, Konrad PE, Dawant BM (2010) CranialVault and its CRAVE tools: a clinical computer assistance system for deep brain stimulation (DBS) therapy. Med Image Anal. doi:10.1016/j.media.2010.07.009
Guo T, Finnis KW, Parrent AG, Peters TM (2006) Visualization and navigation system development and application for stereotactic deep-brain neurosurgeries. Comput Aided Surg 11: 231–239
Guo T, Parrent AG, Peters TM (2007) Surgical targeting accuracy analysis of six methods for subthalamic nucleus deep brain stimulation. Comput Aided Surg 12: 325–334
Collins DL, Holmes CJ, Peters T, Evans AC (1995) Automatic 3-D model based neuroanatomical segmentation. Hum Brain Mapp 3: 190–208
Fonov V, Evans AC, Botteron K, Almli CR, McKinstry RC, Collins DL, BrainDevelopment Cooperative Group (2011) Unbiased average age-appropriate atlases for pediatric studies. Neuroimage 54: 313–327
Avants BB, Epstein CL, Grossman M, Gee JC (2008) Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med Image Anal 12(1): 26–41
Coupé P, Manjon JV, Fonov V, Pruessner J, Robles M, Collins DL (2011) Patch-based segmentation using expert priors: application to hippocampus and ventricle segmentation. Neuroimage 54: 940–954
Neelin P (1998) The MINC file format:from bytes to brains. NeuroImage 7(4): 786
Sled JG, Zijdenbos AP, Evans AC (1998) A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imaging 17: 87–97
Nyul LG, Udupa JK, Saha PK (2003) Incorporating a measure of local scale in voxel-based 3-D image registration. IEEE Trans Med Imaging 22: 228–237
Wells WM III, Viola P, Atsumi H, Nakajima S, Kikinis R (1994) Multi-modal volume registration by maximization of mutual information. Med Image Anal 1(1): 35–51
Collins DL, Neelin P, Peters TM, Evans AC (1994) Automatic 3D intersubject registration of MR volumetric data in standardized Talairach space. J Comput Assist Tomogr 18: 192–205
Smith SM (1998) Fast robust automated brain extraction. Hum Brain Mapp 17: 143–155
Mai JK, Paxinos G, Voss T (2008) Atlas of the human brain. Elsevier Inc, Amsterdam
Coupé P, Yger P, Prima S, Hellier P, Kervrann C, Barillot C (2008) An optimized blockwise nonlocal means denoising filter for 3-D magnetic resonance images. IEEE Trans Med Imaging 27: 425–441
Zijdenbos AP, Dawant BM, Margolin RA, Palmer AC (1994) Morphometric analysis of white matter lesions in MR images: method and validation. IEEE Trans Imaging 13: 716–724
Dice L (1945) Measure of the amount of ecological association between species. Ecology 26(3): 297–302
Hu S, Coupe P, Pruessner JC, Collins DL (2011) Appearance-based modeling for segmentation of hippocampus and amygdala using multi-contrast MR imaging. Neuroimage 58: 549–559
Bhattachargee M, Pitiot A, Roche A, Dormont D, Bardinet E (2008) Anatomy-preserving nonlinear registration of deep brain ROIs using confidence-based block-matching. In: LNCS MICCAI, part II, Springer, vol 5242, pp 956–963. doi:10.1007/978-3-540-85990-1_115
Bardinet E, Dormont D, Malandain G, Bhattachargee M, Pidoux B, Saleh C, Cornu P, Ayache N, Agid Y, Yelnik J (2005) Retrospective cross-evaluation of an histological and deformable 3D atlas of the basal ganglia on series of Parkinsonian patients treated by deep brain stimulation. In: LNCS MICCAI, Springer, vol 3750, pp 385–393. doi:10.11007/11566489_48
Chakravarty MM, Sadikot AF, Germann J, Hellier P, Bertrand G, Collins DL (2009) Comparison of piece-wise linear, linear, and nonlinear atlas-to-patient warping techniques: analysis of the labelling of subcortical nuclei for functional neurosurgical applications. Hum Brain Mapp 30: 3574–3595
Klein A, Andersson J, Ardekani BA, Ashburner J, Avants B, Chiang MC, Christensen GE, Collins DL, Gee J, Hellier P, Song JH, Jenkinson M, Lepage C, Rueckert D, Thompson P, Vercauteren T, Woods RP, Mann JJ, Parsey RV (2009) Evaluation of 14 nonlinear deformation algorithms applied to human brain MRI registration. Neuroimage 46: 786–802
Collins DL, Pruessner JC (2010) Towards accurate, automatic segmentation of the hippocampus and amygdala from MRI by augmenting ANIMAL with a template library and label fusion. Neuroimage 52: 1355–1366
Aljabar P, Heckeman RA, Hammers A, Hajnal JV, Rueckert D (2009) Multi-atlas based segmentation of brain images: atlas selection and its effect on accuracy. Neuroimage 46: 726–738
Patenaude B, Smith SM, Kennedy DN, Jenkinson M (2011) A Bayesian model of shape and appearance for subcortical brain segmentation. Neuroimage 56: 907–922
Antonini A, Isaias IU, Rodolfi G, Landi A, Natuzzi F, Siri C, Pezzoli G (2011) A 5-year prospective assessment of advanced Parkinson disease patients treated with subcutaneous apomor- phine infusion or deep brain stimulation. J Neurol 258: 579–585. doi:10.1007/s00415-010-5793-z
Merola A, Zibettu M, Angrisano S, Rizzi L, Lanotte M, Lopiano L (2011) Comparison of subthalamic nucleus deep brain stimulation and duodopa in the treatment of advanced Parkinson’s disease. Mov Disord 26(4): 664–670
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Haegelen, C., Coupé, P., Fonov, V. et al. Automated segmentation of basal ganglia and deep brain structures in MRI of Parkinson’s disease. Int J CARS 8, 99–110 (2013). https://doi.org/10.1007/s11548-012-0675-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-012-0675-8