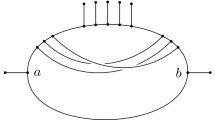
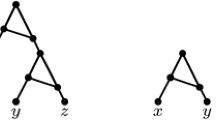
Abstract
Internodons are a formalization of Hennig’s concept of species. We present an alternative construction of internodons imposing a tree structure on the genealogical network. We prove that the segments (trivial unary trees) from this tree structure are precisely the internodons. We obtain the following spin-offs. First, the generated tree turns out to be an organismal tree of life. Second, this organismal tree is homeomorphic to the phylogenetic Hennigian species tree of life, implying the discovery of a multi-level tree of life: this phylogenetic tree can be obtained by zooming out from the organismal tree, or conversely, the organismal tree of life can be generated by expanding the phylogenetic nodes into unary trees. Finally, the definition of the organismal tree allows an efficient algorithmic transformation of a given genealogical network into its corresponding phylogenetic species tree of life. The latter will be presented in a separate paper.
















Similar content being viewed by others
Notes
“If, therefore, the relationships between the elements of a hierarchy are represented by unidirectional arrows, then according to Woodger’s definition: (1) The point of one, and only one, arrow can lie in each element of the hierarchy, whereas several arrows may arise from it. (2) There is one, and only one, element from which arrows emanate but to which no arrows lead. Woodger and Gregg call this element the “beginner”. (3) All elements to which an arrow leads, and which therefore lie at an arrow tip, are connected with the beginner by an arrow or a sequence of arrows.” Hennig (1966: p. 17)
This practice fits into ‘stem-based tree’ terminology, not to be confused with ‘node-based tree’ terminology where the nodes represent the species. See Martin et al. (2010) and Podani (2013) for discussion of mixing node- and stem-based terminology. Confusion about the nodal part of the name could be avoided by calling it the ‘intersplittal species concept.’
In hindsight perhaps better named “intersplittons.”
That is, information pertaining to the parenthood relation between organisms (as defined by the genealogical network).
It would be more appropriate, but less natural, to speak of birthmoments rather than birthdates. We impose the constraint that no two members of the population have the same birthdate: a constraint that makes more sense the finer we divide time. Formally, the role birthdates play is to extend the partial order of ancestorhood into a total order, and for this purpose, we need not worry about the precise details of what exactly defines an organism’s exact moment of birth.
If, in clause 1 of this definition, we would have stipulated birthdates to be natural numbers instead of reals, then downward finiteness would follow immediately from uniqueness of birthdates. In the sequel of this paper, it will transpire that birthdates are needed only insofar as they impose a topological order on the graph, and natural numbers are sufficient for this purpose. However, we chose to stick to defining birthdates as real numbers because this is closer to the day-to-day meaning of this notion.
Here, \(\frown \) denotes concatenation and \(\rho ^{{\mathrm {reverse}}}\) stands for the path \(\rho \) travelled in the opposite direction. More formally, if \(\rho =a_1,\ldots ,a_n\) denotes an undirected path (where the \(a_i\) are nodes), then \(\rho ^{{\mathrm {reverse}}}\) is the path \(a_n,\ldots ,a_1\).
See Velasco (2008) for more discussion on the problem of short-lived internodons.
Since \(p_i\) and \(p_{i+1}\) are consecutive vertices on an undirected path in \(G\), they are adjacent in \(G\). So if \(p_i\) is not a parent of \(p_{i+1}\), then \(p_i\) must be a child of \(p_{i+1}\).
Thus, for any two SD vertices \(x,y\), if \(x\) is an undirparent of \(y\), then there is an arc from \(x\) to \(y\) in \(G'_{\mathbf {SD}}\).
Note that a non-final vertex in a segment of \(G'_{\mathbf {SD}}\) may have outdegree \(>1\) in \(G'\), but must have exactly one SD undirchild.
Note that \(y\in I\) implies that \(y\) is SD, which implication is also true if \(x{\mathbf {INTSD}}y\). Thus, the Proposition would not change if we wrote “for any SD organism \(y\).”
References
Alexander S (2013) Infinite graphs in systematic biology, with an application to the species problem. Acta Biotheor 61:181–201. http://arxiv.org/abs/1201.2869
Darwin C (1872) On the origin of species, 6th edn. John Murray, London
Darwin F (1898) The Knight-Darwin law. Nature 58:630–632
Dress A, Moulton V, Steel M, Wu T (2010) Species, clusters and the ‘Tree of life’: a graph-theoretic perspective. J Theor Biol 265:535–542. http://arxiv.org/abs/0908.2885
Hennig W (1966, reprinted 1979) Phylogenetic systematics. (trans: Davis D, Zangerl R). University of Illinois Press, Urbana IL.
Kahn A (1962) Topological sorting of large networks. Comm ACM 5:558–562
Kornet DJ (1993) Permanent splits as speciation events: a formal reconstruction of the internodal species concept. J Theor Biol 164:407–435
Kornet DJ, Metz JAJ, Schellinx HAJM (1995) Internodons as equivalence classes in genealogical networks: building-blocks for a rigorous species concept. J Math Bio 34:110–122
Kornet DJ, McAllister J (2005) The composite species concept: a rigorous basis for cladistic practice. In: Reydon T, Hemerik L (eds) Current themes in theoretical biology: a dutch perspective. Springer, Dordrecht, pp 95–127
Martin J, Blackburn D, Wiley EO (2010) Are node-based and stem-based clades equivalent? insights from graph theory. PLOS Curr 2:1–12
Nixon KC, Wheeler QD (1990) An amplification of the phylogenetic species concept. Cladistics 6:211–223
Podani J (2013) Tree thinking, time and topology: comments on the interpretation of tree diagrams in evolutionary/phylogenetic systematics. Cladistics 29(3):315–327
Velasco J (2008) The internodal species concept: a response to ‘The tree, the network, and the species’. Biol J Linn Soc 93:865–869
Woodger JH (1952) From biology to mathematics. Br J Philos Sci 3:1–21
Acknowledgments
We thank Rino Zandee for inspiring comments and profitable discussions. We thank the anonymous reviewers and the handling editor for their constructive remarks that helped us improve the readability of the paper.
Figures 1, 2, 7 and 9 have been reproduced from Infinispec (www.zandee.net/infinispec), copyright 2013 by the license holders, with their kind permission.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Alexander, S.A., de Bruin, A. & Kornet, D.J. An Alternative Construction of Internodons: The Emergence of a Multi-level Tree of Life. Bull Math Biol 77, 23–45 (2015). https://doi.org/10.1007/s11538-014-0048-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11538-014-0048-2
Keywords
- Hennigian species
- Internodon
- Organismal tree of life
- Undirparent relation
- Species algorithms
- Multi-level tree of life
- Genealogical network transformation