Abstract
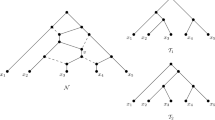
In the last decade, the use of phylogenetic networks to analyze the evolution of species whose past is likely to include reticulation events, such as horizontal gene transfer or hybridization, has gained popularity among evolutionary biologists. Nevertheless, the evolution of a particular gene can generally be described without reticulation events and therefore be represented by a phylogenetic tree. While this is not in contrast to each other, it places emphasis on the necessity of algorithms that analyze and summarize the tree-like information that is contained in a phylogenetic network. We contribute to the toolbox of such algorithms by investigating the question of whether or not a phylogenetic network embeds a tree twice and give a quadratic-time algorithm to solve this problem for a class of networks that is more general than tree-child networks.





Similar content being viewed by others
References
Cardona G, Llabrés M, Rosselló F, Valiente G (2008) A distance metric for a class of tree-sibling phylogenetic networks. Bioinformatics 24:1481–1488
Cardona G, Rossello F, Valiente G (2009) Comparison of tree-child phylogenetic networks. IEEE Trans Comput Biol Bioinform 6:552–569
Cormen TH, Leiserson CE, Rivest RL, Stein C (2001) Introduction to algorithms. MIT Press and McGraw-Hill, New York
Huson DH, Rupp R, Scornavacca C (2010) Phylogenetic networks: concepts, algorithms and applications. Cambridge University Press, Cambridge, MA
van Iersel L, Semple C, Steel M (2010) Locating a tree in a phylogenetic network. Inf Process Lett 110:1037–1043
Kanj IA, Nakhleh L, Than C, Xia G (2008) Seeing the trees and their branches in the network is hard. Theor Comput Sci 401:153–164
Linz S, St. John K, Semple C (2013) Counting trees in a phylogenetic network is #P-complete. SIAM J Comput 42:1768–1776
McDiarmid C, Semple C, Welsh D (in press) Counting phylogenetic networks. Ann Comb
Nakhleh L, Jin G, Zhao F, Mellor-Crummey J (2005) Reconstructing phylogenetic networks using maximum parsimony. In: IEEE computational systems bioinformatics conference, pp 440–442
Willson SJ (2010) Properties of normal phylogenetic networks. Bull Math Biol 72:340–358
Willson SJ (2012) Tree-average distances on certain phylogenetic networks have their weights uniquely determined. Algorithm Mol Biol 7:13
Acknowledgments
We thank the two anonymous referees for their helpful comments.
Author information
Authors and Affiliations
Corresponding author
Additional information
We thank the Allan Wilson Centre for Molecular Ecology and Evolution, the New Zealand Marsden Fund, and the 7th European Community Framework Programme for their financial support.
Rights and permissions
About this article
Cite this article
Cordue, P., Linz, S. & Semple, C. Phylogenetic Networks that Display a Tree Twice. Bull Math Biol 76, 2664–2679 (2014). https://doi.org/10.1007/s11538-014-0032-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11538-014-0032-x