Abstract
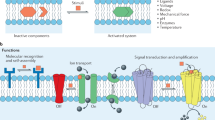
Cell membranes exhibit multiple layers of complexity, ranging from their specific molecular content to their emergent mechanical properties and dynamic spatial organization. Both compositional and geometrical organizations of membrane components are known to play important roles in life processes, including signal transduction. Supported membranes, comprised of a bilayer assembly of phospholipids on the solid substrate, have been productively served as model systems to study wide range problems in cell biology. Because lateral mobility of membrane components is readily preserved, supported lipid membranes with signaling molecules can be utilized to effectively trigger various intercellular reactions. The spatial organization and mechanical deformation of supported membranes can also be manipulated by patterning underlying substrates with modern micro- and nano-fabrication techniques. This article focuses on various applications and methods to spatially patterned biomembranes by means of curvature modulations and spatial reorganizations, and utilizing them to interface with live cells. The integration of biological components into synthetic devices provides a unique approach to investigate molecular mechanisms in cell biology.
Similar content being viewed by others
1 Introduction
In biology, the lipid bilayer membrane was first identified as the barrier defining the interior and exterior of cells. As our understanding of cell membranes developed over the years, it has become clear that the complex liquid interface provided by membranes also serves as a primary platform for cellular signal transduction machinery. Indeed, nearly all molecular life processes involve membranes at some point. The basic structural component of cell membranes, the lipid bilayer, has been known to be a two-dimensional fluid since the 1970s [46]. Rather than a homogeneous fluid sheet, lipid membranes in the cell exhibit heterogeneity and flexibility both in compositions and organizations [35]. Now, the spatial organization and morphology of cellular membranes and membrane components are widely known to mediate numerous biological functions [12, 21, 30, 51, 52].
The cellular membrane creates an environment with many unique characteristics for both self-assembled and actively driven molecular sorting processes. Supported lipid membranes, formed by spontaneous fusion of small lipid vesicles onto a hydrophilic surface such as silicon oxide [3], exhibits similar two-dimensional fluidity in vitro as natural cell membranes and has been broadly utilized as a model system to study membrane organizations [4, 13, 21, 29, 31, 39, 42, 49, 50]. Lipids and membrane-linked proteins in the supported membrane can diffuse freely over macroscopic distances with lateral diffusion coefficients generally on the order of 1 μm2/s [22, 43, 48]. As the chemical composition of supported membranes can be easily manipulated, the model membrane system creates many possibilities to represent biocompatible interfaces with controllable physical properties, such as electrostatic charges [17, 28], surface density of molecules [29, 40], and degree of molecule clustering [1, 23].
Another salient characteristic of the support membrane is its compatibility to merge diverse research techniques, such as micro/nano-fabrication and fluorescence microscopy. Solid supported membrane substrates can be spatially patterned via modern techniques [22, 27, 34, 39, 45]. By fabricating barriers on the substrate, typically thin metal lines, supported membranes can be partitioned into different regions while membrane components are locally confined. Direct membrane patterning can also be achieved by UV/ozone photolithography which directly removes lipid membrane [47, 58]. Concurrently, underlying substrates can be fabricated with three-dimensional features and create curvature modulation of supported membranes [42, 56]. Combined with biocompatible surface chemistry techniques, signaling molecules can be represented along with patterned supported membranes, too. Spatial and curvature-patterned supported membranes readily provide distinctive experimental setup to investigate many research aspects in cell biology, such as immunology and cancer biology.
2 Supported membranes and immunological synapses
The immunological synapse between T cell and antigen-presenting cell (APC) involves large scale spatial sorting of membrane proteins in concert with intracellular T cell signaling activity. Previously, functionalized supported membranes have been demonstrated to effectively trigger T cell activations and form hybrid immunological synapses. APC membrane proteins, including antigenic peptide loaded major histocompatibility complex II (pMHC) and intercellular adhesion molecule 1 (ICAM-1) can be chemically attached to lipid membranes and preserve two-dimensional mobility. As the T cell is engaged, T cell receptor (TCR) ligates with mobile pMHC and together form central supramolecular activation complex (c-SMAC) in the immunological synapse. Although biochemical interactions between molecules in immunological synapses have been extensively studied for many years, hybrid immunological synapses provide unique advantages in the study of spatial redistribution of membrane proteins in live T cells.
The lateral mobility of molecules in the supported membrane can be controlled by barriers fabricated onto the underlying substrate. Nanometer scale metal grids, such as thin chromium lines, physically partition supported membranes into separated membrane corrals. Molecules diffuse and assemble normally within each corral but cannot hop across the barriers. When these patterned supported membranes engage with living cells, cell surface receptors whose cognate ligands are in the supported membrane become subject to the same constraints on molecular movement. In turn, other signaling molecules inside the cell that associate with these receptors also follow the prescribed organization. In this way, it is possible to control the spatial organization of the signal transduction machinery for a specific signaling pathway in living cells. We have referred to such manipulations as spatial mutations; essentially chemically identical cells differing only by their molecular spatial organization are created.
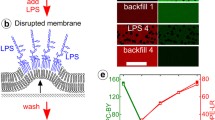
The spatial mutation strategy has proven useful in studies of molecular spatial organization within immunological synapses (Fig. 1). For example, during immunological synapse formation, T cell receptors (TCR) first bind to their cognate ligands, antigenic peptide MHC, and assemble into microclusters. The formation of these microclusters is critical to TCR signaling and, unlike solid substrate display strategies [11], supported membranes allow this type of clustering to occur. At sufficiently high antigen level, TCR microclusters are subsequently transported radially towards the junction center resulting in a bull’s-eye pattern with TCR-MHC in the middle and other adhesion molecules forming peripheral rings. It is this pattern that was first named the immunological synapse [18]. Naturally, there was much interest in the roll, if any, such a large scale pattern had in signal transduction processes. Using the spatial mutation method in its debut, Mossman et al. [39] demonstrated that TCR clusters were indeed signaling differently if they were physically trapped in the periphery of the cell. Since this first introduction, various forms of the technology have been applied to other aspects of the T cell synapse [9, 57].
Spatial patterning of fluid supported membranes and applications in live T cell immunological synapses. Various methods have been developed to locally confine two-dimensional mobility in supported membranes. Here, passive grid barriers on the glass support are fabricated by electron-beam lithography and provide lateral confinement of membrane components. By fluorescence microscopy, the bull’s-eye protein pattern of the immunological synapse in live T cell is altered by the presence of grid barriers on supported membrane (right panel). Scale bar 5 μm
Another type of supported membrane patterning has been achieved in the context of the immunological synapse using photo-releasable agonist peptides. In this case, the identity of the agonist peptide, to which the T cell will respond, is masked with a photosensitive chemical moiety 6-nitroveratryloxycarbonyl (NVOC). Irradiations with near-UV light restore agonist activities by photochemically cleaving NVOC protecting group [10, 25]. The ability to photoactivate protein functions on the supported membrane enables both spatial and temporal manipulations to investigate underlying mechanisms of immune synapse assembly and signaling.
3 Other juxtacrine signaling: EphA2–EphrinA1 in cancer biology
Another signaling system that shares the juxtacrine configuration of receptors on one cell membrane with their membrane-bound ligands on an apposed cell membrane is EphA2–EphrinA1. EphA2 is a receptor tyrosine kinase and is a member of the large family of Eph receptors, which collectively govern many aspects of cellular assembly into tissues [33, 38]. Upon ligation with endogenous EphrinA1 on the plasma membrane of apposed cells, activated EphA2 can initiate various signal pathways and misregulation of this these pathways contributes to cancer metastasis. It has been reported that naturally monomeric EphrinA1 in solution can bind to EphA2 receptors, but fails to trigger EphA2 signal cascades unless it is artificially linked into dimers [8]. In spite of this shortfall, artificial EphrinA1 dimers have been widely used to study the EphA2 signaling system.
Display of EphrinA1 on supported membranes provides a great experimental platform to study the Eph signaling with live cells. Notably, mobile EphrinA1 on supported membranes can effectively trigger EphA2 activation in human breast cancer cells. The lateral mobility of EphrinA1 on supported membranes also reveals dynamic reorganizations of EphA2/EphrinA1 clusters during signaling. While the underlying mechanism of EphA2/EphrinA2 macroscopic protein pattern formation is still unclear, application of the spatial mutation technology to this system has enabled investigation the role of spatial redistribution in signaling. Recently reported results unveil a previously unreported mechanical aspect of EphA2 signaling. Specifically, physical restriction of EphA2–EphrinA1 cluster movement by substrate barriers alters the cellular response to EphrinA1 as observed by cytoskeleton reorganization and protease recruitment [44]. In light of EphA2’s role in cancer and the known mechanical changes to tissue in cancer, these patterned membrane techniques have helped to reveal a possible molecular mechanism by which tissue mechanics contributes to cancer progression.
4 Techniques for membrane patterning
Patterned supported membrane, by means of controlling spatial organization, chemical functionalization, curvature, and topography, provides numerous opportunities to probe and control living cells. Various techniques for membrane patterning have been developed since last decade and we present a brief overview of the state of the art.
5 Spatial patterning
Nano/micro-fabricated lines of materials that resist membrane formation (e.g. metals, some polymers, and proteins) can serve as passive barriers to molecular lateral mobility in the membrane as discussed above. High-resolution patterns on underlying substrates are achieved by various modern solid-state fabrication techniques, including photolithography, electron-beam lithography, and thin film deposition [9, 26, 39]. Typically, thin chromium lines with 100 nm in width and 5 nm in height are fabricated on the glass substrate. As lipid molecules cannot self-assemble a continuous bilayer membrane over chromium surface, the nanopatterned metal grids on the glass substrate effectively act as barriers for membrane components.
Direct removal of lipid molecules and proteins has also been demonstrated to pattern supported membranes. UV/ozone photolithography, for example, can locally degrade and remove lipids and proteins from supported membranes and the resultant patterns remain intact [55, 58]. High resolution of in situ lipid removal can be achieved by multiphoton photolithography using near-infrared Ti:sapphire laser and high NA lens on the microscope [47]. Scanning probe lithography can also be utilized to pattern supported membranes. Using scanning tips of atomic force microscope, lipid molecules can be physically removed from bilayer structures [26, 45]. Patterns generated by direct lipid removal can be preserved by back-filling with blocking proteins, such as bovine serum albumin. Or, when performed on substrates prepatterned with grid lines, depleted corrals can be refilled with another composition of fluid membrane [26].
Lift-off strategies have been implemented to pattern supported membrane as well. A sacrificial layer, such as thin aluminum metal, is prepatterned on the glass substrate. After supported membrane formation, underlying aluminum layer are then etched in mildly basic aqueous solution and lipid molecules over aluminum region are removed [27]. Similarly, prepatterned parylene polymer on the glass substrate can also be lifted off and then create complementarily patterned supported membranes [51].
Other patterning techniques with lower spatial resolutions have also been used in other biological applications. For example, supported membranes in microfluidics channels combine both advantages of small volume in microfluidics and lateral mobility of membrane components and have been greatly utilized as ligand detection systems in vitro [29]. Direct microcontact printing of lipid molecules by plasma-oxidized polydimethylsiloxane (PDMS) stamps have been used to create array patterns of supported membranes [54]. Large scale supported membrane arrays can be printed by microarrayer under humidified conditions and have industrial applications [53].
6 Curvature patterning
Cellular membranes display a variety of morphology with different curvatures. It is believed that membrane geometry could be dynamically regulated in order to provide appropriate environment and to perform various biological processes, including vesicle secretion, motility, and endocytosis. Therefore, controlling membrane curvature is important to study the coupling between membrane deformation and signal transduction [20, 37].
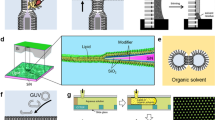
With supported membranes, the modulation of membrane deformation can be accomplished by three-dimensional microfabrication on the underlying substrate [42]. After photolithography, glass substrates are fabricated by combinations of anisotropic and isotropic etching processes. Various substrate curvatures can be achieved by controlling etching processes. Actual surface profile of patterned substrates can then be characterized by atomic force microscopy, for example, to provide high-resolution information. A continuous supported lipid membrane can then deposit on the curvature-patterned substrates by spontaneous fusion of small lipid vesicles.
In the study of cholesterol-dependent phase-separation, controlling curvature in supported membrane has been successfully utilized to spatially organize phase-separated membrane domains [42]. Recently, this membrane curvature modulation technique has been applied to mechanically perturb immunological synapses in live T cells. Initial observation indicates that large scale of protein patterns in the immunological synapse can be altered merely by imposing a defined membrane curvature (Fig. 2). This mechanical perturbation via curvature modulations also changes overall cell morphology. The curvature-patterned hybrid cell junctions surely provide new strategies to study the effect of membrane curvature modulation in cellular signal transduction, such as in cytoskeletal regulations.
Curvature patterning of supported membranes and applications in live T cell immunological synapses. a Schematic of imposing membrane curvature onto immunological synapses in live T cells. The modulation of membrane Curvature is accomplished by three-dimensional microfabrication on the underlying substrate. b, c Curvature-patterned immunological synapses is visualized by reflection interference contrast microscopy (b, RICM) and fluorescence microscopy (c). The morphological changes in cell contact area, generally with lower intensity duo to destructive interference in RICM, indicated that membrane curvature plays an important role in cellular responses. Canonical bull’s-eye protein pattern of TCR in immunological synapse is distorted merely by imposing membrane deformations. Scale bar 10 μm
7 Biological functionalization
For biological studies, membrane functionalization generally requires the attachment of specific proteins to the lipid bilayer. As supported membranes require aqueous environment to maintain the lipid bilayer structure, a variety of specialized biocompatible techniques have been developed to functionalize supported membranes (summarized in Table 1).
Lipid molecules with nitrilotriacetic acid in the head group (NTA lipids) provide stable anchor points using chelation chemistry (Fig. 3). Under neutral pH, proteins which have histidine tags expressed can be chelated with NTA lipids loaded with Ni2+ ion[40]. Multiple histidine residues can be sequentially added and ensure stable attachments and preserve proper orientations over supported membranes. In general, 1–5 mol% of 1,2-dioleoyl-sn-glycero-3-[(N-(5-amino-1-carboxypentyl)iminodiacetic acid)succinyl] NTA-DOGS lipids are doped into supported membranes. Ni–NTA lipid–protein linkages have been successfully utilized in various biological researches, such as T cell immunological synapse describe above.
Biological functionalization of supported membranes. Various chemical attachment schemes can be utilized to link singling molecules onto supported membranes. a Proteins with histidine residues can be chelated with Ni2+ loaded NTA-lipids on supported membranes. Multiple exposed histidine residues, such as His6 or His10, can ensure stably attachment within 24 hours. b Covalent attachment via maleimide and thiol reactions. Proteins with exposed thiol can be covalently linked to maleimide-lipids. c Molecules with natural or synthetic glycosylphosphatidylinositol (GPI) tails can be directly incorporated into supported membranes through the insertion of GPI tails into lipid bilayer structures
Alternately, lipids with maleimide head group (maleimide lipid) can be doped into supported membranes and allow covalent linkage to thiol group. 1,2-Dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-[4-(p-maleimidomethyl)cyclohexane-carboxamide] (MCC-PE) is commonly added in supported membranes with 1–10 mol% concentration. Molecules with exposed thiol group, such as cysteine residues in proteins, can be covalently linked to maleimide lipid in mild basic conditions (above pH 8) [14]. Irreversible and stable attachment of maleimide–thiol lipid–protein linkages have been employed to study kinetics of membrane-dependent activation of Ras signaling systems.
Natural or synthetic glycosylphosphatidylinositol (GPI) linkage is another attachment strategy to supported membranes. Proteins with exposed GPI can be incorporated into supported membranes through the insertion of GPI tails into lipid bilayer structures [18]. This technique was widely used in earlier studies of supported membranes with cells, but has more recently been supplanted by easier linkage systems (especially multiple histidine chelation). One problem with GPI linkages is that they require cell membrane preparations and membrane protein purifications, and thus are much more cumbersome. A more insidious problem with GPI linkages is some evidence for purified GPI-linked proteins to form unnatural aggregations in membranes. Other attachment schemes, such as stable binding between biotin and streptavidin is widely utilized because of its ease and availability. Biotinylated lipid can be doped into supported membranes, and streptavidin can serve as a stable linking bridge between both biotinylated target and lipid molecules [31]. This is a reasonable starting point for membrane functionalization, but it is important to realize that streptavidin introduces a large amount of oligomerization into the system, which could lead to side effects in some situations.
Recently, some strategies to combine immobilized proteins (in controlled patterns) within mobile supported membranes are emerging. Covalent immobilizations are usually achieved by using heterobifunctional crosslinkers [24]. One functional group in crosslinkers selectively reacts with micro- or nano-patterned substrate, such as silanization over glass and self-assemble monolayer over gold. The other reactive group in crosslinkers, such as N-hydroxysulfosuccinimide ester, maleimide, and carbodiimide analog, biochemically reacts with target proteins at primary amine, thiol, and carboxyl groups, respectively. Since immobilized molecules inherently serve as diffusion barriers, the combined representation of immobile and mobile components in supported membranes create new possibilities to study membrane signal transduction in cell biology.
8 Polymer-tethered lipid membranes
While lipid membranes on glass substrates create various research opportunities in cell biology, polymer-supported lipid membranes allow ligand integrations even with native transmembrane domains [1, 50]. Phospholipids attached with polyethylene glycol (PEG) polymer are often utilized as physical cushions to keep transmembrane molecules from nonspecific interactions with supporting substrates. Polymer-supported lipid membranes are proven useful in recent studies of vesicle fusions with molecule complexes of soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNARE) in vitro [32]. Currently, spatial-patterning techniques and integration of in vivo cell biological researches using polymer-supported lipid membranes are mostly under development. Nevertheless, polymer-tethered lipid membranes may offer unique solutions to answer certain types of biological questions.
9 Additional applications and future directions
In addition to cell–cell signaling, supported lipid membrane technologies facilitate many biological applications, including lipid membrane arrays for drug discovery [19, 53] and patch clamps for electrophysiology [36]. With advanced optical microscopy techniques, molecular orientations of membrane-associated molecules, such as mucin glycoproteins can be measured with nanometer resolution on supported lipid membranes.[16, 41] Supported membranes with different charged lipid molecules can also be utilized to modulate electrostatic potential on surfaces and provide alternate strategies for molecule detections[2, 7]. Recently, integration of supported membranes with plasmonic nanocubes effectively forms a label-free biosensor that measures protein binding to lipid membranes [15].
As numerous membrane functionalization techniques have been introduced, orthogonal linking schemes in identical chemical conditions would potentially expand multi-component representations on supported lipid membranes and further mimic the complexity in cellular interfaces. Because of the high selectivity and sensitivity, complementary single-strand DNA hybridization may serve as stable linking schemes between lipid molecules and proteins [5, 6]. Patterned supported membrane technology began to emerge more than a decade ago, and the field has experienced significant growth over the years. Perhaps most importantly, applications of this technology to real problems in cell biology are also developing. Looking forward, we anticipate hand-in-hand growth of both technical capabilities and cell biological applications of patterned membrane systems.
References
Albertorio F, Diaz AJ, Yang T, Chapa VA, Kataoka S, Castellana ET, Cremer PS (2005) Fluid and air-stable lipopolymer membranes for biosensor applications. Langmuir 21:7476–7482
Baksh MM, Jaros M, Groves JT (2004) Detection of molecular interactions at membrane surfaces through colloid phase transitions. Nature 427:139–141
Brian AA, McConnell HM (1984) Allogeneic stimulation of cytotoxic T cells by supported planar membranes. Proc Natl Acad Sci USA 81:6159–6163
Chan YH, Boxer SG (2007) Model membrane systems and their applications. Curr Opin Chem Biol 11:581–587
Chan YH, van Lengerich B, Boxer SG (2009) Effects of linker sequences on vesicle fusion mediated by lipid-anchored DNA oligonucleotides. Proc Natl Acad Sci USA 106:979–984
Chung M, Lowe RD, Chan YH, Ganesan PV, Boxer SG (2009) DNA-tethered membranes formed by giant vesicle rupture. J Struct Biol 168:190–199
Clack NG, Salaita K, Groves JT (2008) Electrostatic readout of DNA microarrays with charged microspheres. Nat Biotechnol 26:825–830
Davis S, Gale NW, Aldrich TH, Maisonpierre PC, Lhotak V, Pawson T, Goldfarb M, Yancopoulos GD (1994) Ligands for EPH-related receptor tyrosine kinases that require membrane attachment or clustering for activity. Science 266:816–819 (New York, NY )
DeMond AL, Mossman KD, Starr T, Dustin ML, Groves JT (2008) T cell receptor microcluster transport through molecular mazes reveals mechanism of translocation. Biophys J 94:3286–3292
DeMond AL, Starr T, Dustin ML, Groves JT (2006) Control of antigen presentation with a photoreleasable agonist peptide. J Am Chem Soc 128:15354–15355
Doh J, Irvine DJ (2006) Immunological synapse arrays: patterned protein surfaces that modulate immunological synapse structure formation in T cells. Proc Natl Acad Sci USA 103:5700–5705
Dustin ML (2009) Supported bilayers at the vanguard of immune cell activation studies. J Struct Biol 168:152–160
Dustin ML, Starr T, Varma R, Thomas VK (2007) Supported planar bilayers for study of the immunological synapse. Coligan JE et al (eds) Current protocols in immunology, chapter 18, unit 18 13
Galush WJ, Nye JA, Groves JT (2008) Quantitative fluorescence microscopy using supported lipid bilayer standards. Biophys J 95:2512–2519
Galush WJ, Shelby SA, Mulvihill MJ, Tao A, Yang P, Groves JT (2009) A nanocube plasmonic sensor for molecular binding on membrane surfaces. Nano letters 9:2077–2082
Godula K, Umbel ML, Rabuka D, Botyanszki Z, Bertozzi CR, Parthasarathy R (2009) Control of the molecular orientation of membrane-anchored biomimetic glycopolymers. J Am Chem Soc 131:10263–10268
Gomez EW, Clack NG, Wu H-J, Groves JT (2009) Like-charge interactions between colloidal particles are asymmetric with respect to sign. Soft Matter 5:1931–1936
Grakoui A, Bromley SK, Sumen C, Davis MM, Shaw AS, Allen PM, Dustin ML (1999) The immunological synapse: a molecular machine controlling T cell activation. Science (New York, NY) 285:221–227
Groves JT (2002) Membrane array technology for drug discovery. Curr Opin Drug Discov Dev 5:606–612
Groves JT (2007) Bending mechanics and molecular organization in biological membranes. Annu Rev Phys Chem 58:697–717
Groves JT, Dustin ML (2003) Supported planar bilayers in studies on immune cell adhesion and communication. J Immunol Methods 278:19–32
Groves JT, Ulman N, Boxer SG (1997) Micropatterning fluid lipid bilayers on solid supports. Science (New York, NY) 275:651–653
Hartman NC, Nye JA, Groves JT (2009) Cluster size regulates protein sorting in the immunological synapse. Proc Natl Acad Sci USA 106:12729–12734
Hermanson GT (2008) Bioconjugate techniques. Elsevier Academic Press, Amsterdam
Huse M, Klein LO, Girvin AT, Faraj JM, Li QJ, Kuhns MS, Davis MM (2007) Spatial and temporal dynamics of T cell receptor signaling with a photoactivatable agonist. Immunity 27:76–88
Jackson BL, Groves JT (2004) Scanning probe lithography on fluid lipid membranes. J Am Chem Soc 126:13878–13879
Jackson BL, Groves JT (2007) Hybrid protein-lipid patterns from aluminum templates. Langmuir 23:2052–2057
Jackson BL, Nye JA, Groves JT (2008) Electrical manipulation of supported lipid membranes by embedded electrodes. Langmuir 24:6189–6193
Jung H, Robison AD, Cremer PS (2009) Detecting protein-ligand binding on supported bilayers by local pH modulation. J Am Chem Soc 131:1006–1014
Kaizuka Y, Douglass AD, Vardhana S, Dustin ML, Vale RD (2009) The coreceptor CD2 uses plasma membrane microdomains to transduce signals in T cells. J Cell Biol 185:521–534
Kaizuka Y, Douglass AD, Varma R, Dustin ML, Vale RD (2007) Mechanisms for segregating T cell receptor and adhesion molecules during immunological synapse formation in Jurkat T cells. Proc Natl Acad Sci USA 104:20296–20301
Karatekin E, Di Giovanni J, Iborra C, Coleman J, O’Shaughnessy B, Seagar M, Rothman JE (2010) A fast, single-vesicle fusion assay mimics physiological SNARE requirements. Proc Natl Acad Sci USA 107:3517–3521
Kuijper S, Turner CJ, Adams RH (2007) Regulation of angiogenesis by Eph-ephrin interactions. Trends Cardiovasc Med 17:145–151
Lenz P, Ajo-Franklin CM, Boxer SG (2004) Patterned supported lipid bilayers and monolayers on poly(dimethylsiloxane). Langmuir 20:11092–11099
Lingwood D, Simons K (2010) Lipid rafts as a membrane-organizing principle. Science (New York, NY) 327:46–50
Mayer M, Terrettaz S, L G, Stora T, Vogel H (2003) In: Cooper JM, Cass AEG (eds) Biosensors – a practical approach, 2nd edn. Oxford University Press, New York, Oxford pp 153–184
McMahon HT, Gallop JL (2005) Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature 438:590–596
Miao H, Wang B (2009) Eph/ephrin signaling in epithelial development and homeostasis. Int J Biochem Cell Biol 41:762–770
Mossman KD, Campi G, Groves JT, Dustin ML (2005) Altered TCR signaling from geometrically repatterned immunological synapses. Science (New York, NY) 310:1191–1193
Nye JA, Groves JT (2008) Kinetic control of histidine-tagged protein surface density on supported lipid bilayers. Langmuir 24:4145–4149
Parthasarathy R, Rabuka D, Bertozzi CR, Groves JT (2007) Molecular orientation of membrane-anchored mucin glycoprotein mimics. J Phys Chem 111:12133–12135
Parthasarathy R, Yu CH, Groves JT (2006) Curvature-modulated phase separation in lipid bilayer membranes. Langmuir 22:5095–5099
Salafsky J, Groves JT, Boxer SG (1996) Architecture and function of membrane proteins in planar supported bilayers: a study with photosynthetic reaction centers. Biochemistry 35:14773–14781
Salaita K, Nair PM, Petit RS, Neve RM, Das D, Gray JW, Groves JT (2010) Restriction of receptor movement alters cellular response: physical force sensing by EphA2. Science (New York, NY) 327:1380–1385
Shi J, Chen J, Cremer PS (2008) Sub-100 nm patterning of supported bilayers by nanoshaving lithography. J Am Chem Soc 130:2718–2719
Singer SJ, Nicolson GL (1972) The fluid mosaic model of the structure of cell membranes. Science (New York, NY) 175:720–731
Smith AM, Huser T, Parikh AN (2007) Dynamic recompartmentalization of supported lipid bilayers using focused femtosecond laser pulses. J Am Chem Soc 129:2422–2423
Stelzle M, Miehlich R, Sackmann E (1992) Two-dimensional microelectrophoresis in supported lipid bilayers. Biophys J 63:1346–1354
Tamm LK, Groves JT (2009) Supported membranes in structural biology. J Struct Biol 168:1–2
Tanaka M, Sackmann E (2005) Polymer-supported membranes as models of the cell surface. Nature 437:656–663
Torres AJ, Vasudevan L, Holowka D, Baird BA (2008) Focal adhesion proteins connect IgE receptors to the cytoskeleton as revealed by micropatterned ligand arrays. Proc Natl Acad Sci USA 105:17238–17244
Treanor B, Batista FD (2007) Mechanistic insight into lymphocyte activation through quantitative imaging and theoretical modelling. Curr Opin Immunol 19:476–483
Yamazaki V, Sirenko O, Schafer RJ, Nguyen L, Gutsmann T, Brade L, Groves JT (2005) Cell membrane array fabrication and assay technology. BMC Biotechnol 5:18
Yang T, Simanek EE, Cremer P (2000) Creating addressable aqueous microcompartments above solid supported phospholipid bilayers using lithographically patterned poly(dimethylsiloxane) molds. Anal Chem 72:2587–2589
Yee CK, Amweg ML, Parikh AN (2004) Direct photochemical patterning and refunctionalization of supported phospholipid bilayers. J Am Chem Soc 126:13962–13972
Yoon TY, Jeong C, Lee SW, Kim JH, Choi MC, Kim SJ, Kim MW, Lee SD (2006) Topographic control of lipid-raft reconstitution in model membranes. Nat Mater 5:281–285
Yu CH, Groves JT (2010) Altered actin centripetal retrograde flow in physically restricted immunological synapses. PLoS ONE (under review)
Yu CH, Parikh AN, Groves JT (2005) Direct patterning of membrane-derivatized colloids using in situ UV-ozone photolithography. Adv Mater 17:1477–1480
Acknowledgments
C.H.Y. is a research fellow supported by Research Centre of Excellence in Mechanobiology (Singapore) and National Science Council (Taiwan, NSC98-2917-I-564-165). The authors also thank Khalid Salaita, Pradeep Nair, Boryana Manz, Wan-chen Lin, Lars Iverson, and Danny van Noort for their help in manuscript preparations.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Yu, Ch., Groves, J.T. Engineering supported membranes for cell biology. Med Biol Eng Comput 48, 955–963 (2010). https://doi.org/10.1007/s11517-010-0634-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11517-010-0634-x