Abstract
Purpose
Glaciers of the Tianshan Mountains have been geographically isolated for a long time with little human interference. This study aimed to understand the viral community structures and their ecological functions in a unique glacier soil.
Materials and methods
The abundances of purified virus particles and bacteria in the glacier soil were examined using epifluorescence microscopy and quantitative PCR of the 16S rRNA gene, respectively. Metagenomic analysis was employed to investigate the taxonomic and functional compositions as well as the phylogenetic relationship of the functional genes of the viruses in the glacier soil.
Results and discussion
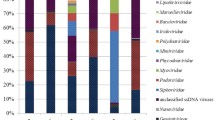
A total of eight double-stranded DNA (dsDNA) virus families were identified in the glacier soil, with Siphoviridae, Podoviridae, and Myoviridae being the more abundant families. The diverse dsDNA viruses and few ssDNA and other types of viruses formed the unique community structure of viruses in the Tianshan Mountains glacier soil. The functional genes identified from the virome mainly belonged to phage-related proteins. The phage terminase of Caudovirales presented high diversity, and its amino acid sequences were different from those in other environments.
Conclusions
The results showed a unique viral diversity and taxonomic composition in the glacier soil, which suggests the significant ecological role of Caudovirales in this environment.



Similar content being viewed by others
References
Adriaenssens EM, Van Zyl L, De Maayer P, Rubagotti E, Rybicki E, Tuffin M, Cowan DA (2015) Metagenomic analysis of the viral community in Namib desert hypoliths. Environ Microbiol 17:480–495
Angly FE, Felts B, Breitbart M, Salamon P, Edwards RA, Carlson C, Chan AM, Haynes M, Kelley S, Liu H, Mahaffy JM, Mueller JE, Nulton J, Olson R, Parsons R, Rayhawk S, Suttle CA, Rohwer F (2006) The marine viromes of four oceanic regions. PLoS Biol 4:2121–2131
Biddle JF, White JR, Teske AP, House CH (2011) Metagenomics of the subsurface Brazos-Trinity Basin (IODP site 1320): comparison with other sediment and pyrosequenced metagenomes. ISME J 5:1038–1047
Black LW (1995) DNA packaging and cutting by phage terminases: control in phage T4 by a synaptic mechanism. BioEssays 17:1025–1030
Breitbart M, Rohwer F (2005) Here a virus, there a virus, everywhere the same virus? Trends Microbiol 13:278–284
Breitbart M, Salamon P, Andresen B, Mahaffy JM, Segall AM, Mead D, Azam F, Rohwer F (2002) Genomic analysis of uncultured marine viral communities. PNAS 99:14250–14255
Breitbart M, Miyake JH, Rohwer F (2004) Global distribution of nearly identical phage-encoded DNA sequences. FEMS Microbiol Lett 236:249–256
Cai L, Zhang R, He Y, Feng X, Jiao N (2016) Metagenomic analysis of virioplankton of the subtropical Jiulong River estuary, China. Viruses 8:35
Cassman N, Prieto-Davó A, Walsh K, Silva GG, Angly F, Akhter S, Barott K, Busch J, McDole T, Haggerty JM (2012) Oxygen minimum zones harbour novel viral communities with low diversity. Environ Microbiol 14:3043–3065
Dell’Anno A, Corinaldesi C, Danovaro R (2015) Virus decomposition provides an important contribution to benthic deep-sea ecosystem functioning. PNAS 112:E2014–E2019
DeLong EF (1992) Archaea in coastal marine environments. PNAS 89:5685–5689
Dinsdale EA, Edwards RA, Hall D, Angly F, Breitbart M, Brulc JM, Furlan M, Desnues C, Haynes M, Li L (2008) Functional metagenomic profiling of nine biomes. Nature 452:629–632
Dong Z, Li Z, Zhang M, Wang F, Wu L (2011) Physico-chemical characteristics and environmental significance of snow deposition on Haxilegen glacier no. 51 in Tian Shan, China. JMS 8:484–494
Han LL, Wang ET, Han TX, Liu J, Sui XH, Chen WF, Chen WX (2009) Unique community structure and biogeography of soybean rhizobia in the saline-alkaline soils of Xinjiang, China. Plant Soil 324:291–305
Hurwitz BL, Westveld AH, Brum JR, Sullivan MB (2014) Modeling ecological drivers in marine viral communities using comparative metagenomics and network analyses. PNAS 111:10714–10719
Jover LF, Effler TC, Buchan A, Wilhelm SW, Weitz JS (2014) The elemental composition of virus particles: implications for marine biogeochemical cycles. Nat Rev Microbiol 12:519–528
Koskella B, Brockhurst MA (2014) Bacteria–phage coevolution as a driver of ecological and evolutionary processes in microbial communities. FEMS Microbiol Rev 38:916–931
Lopez-Bueno A, Tamames J, Velazquez D, Moya A, Quesada A, Alcami A (2009) High diversity of the viral community from an Antarctic Lake. Science 326:858–861
Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y (2012) SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Giga Science 1:1–6
Meyer F, Paarmann D, D’Souza M, Olson R, Glass EM, Kubal M, Paczian T, Rodriguez A, Stevens R, Wilke A (2008) The metagenomics RAST server—a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9:386
Mizuno CM, Rodriguez-Valera F, Kimes NE, Ghai R (2013) Expanding the marine virosphere using metagenomics. PLoS Genet 9:e1003987
Paez-Espino D, Eloe-Fadrosh EA, Pavlopoulos GA, Thomas AD, Huntemann M, Mikhailova N, Rubin E, Ivanova NN, Kyrpides NC (2016) Uncovering Earth’s virome. Nature 536:425–430
Prestel E, Salamitou S, DuBow MS (2008) An examination of the bacteriophages and bacteria of the Namib desert. J Microbiol 46:364–372
Prigent M, Leroy M, Confalonieri F, Dutertre M, DuBow MS (2005) A diversity of bacteriophage forms and genomes can be isolated from the surface sands of the Sahara desert. Extremophiles 9:289–296
Reavy B, Swanson MM, Cock PJ, Dawson L, Freitag TE, Singh BK, Torrance L, Mushegian AR, Taliansky M (2015) Distinct circular single-stranded DNA viruses exist in different soil types. Appl Environ Microbiol 81:3934–3945
Rodriguez-Brito B, Li L, Wegley L, Furlan M, Angly F, Breitbart M, Buchanan J, Desnues C, Dinsdale E, Edwards R (2010) Viral and microbial community dynamics in four aquatic environments. ISME J 4:739–751
Rohwer F, Thurber RV (2009) Viruses manipulate the marine environment. Nature 459:207–212
Roux S, Enault F, Robin A, Ravet V, Personnic S, Theil S, Colombet J, Sime-Ngando T, Debroas D (2012) Assessing the diversity and specificity of two freshwater viral communities through metagenomics. PLoS One 7:e33641
Roux S, Tournayre J, Mahul A, Debroas D, Enault F (2014) Metavir 2: new tools for viral metagenome comparison and assembled virome analysis. BMC Bioinformatics 15:1
Sano E, Carlson S, Wegley L, Rohwer F (2004) Movement of viruses between biomes. Appl Environ Microbiol 70:5842–5846
Short CM, Suttle CA (2005) Nearly identical bacteriophage structural gene sequences are widely distributed in both marine and freshwater environments. Appl Environ Microbiol 71:480–486
Suzuki MT, Taylor LT, DeLong EF (2000) Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5′-nuclease assays. Appl Environ Microbiol 66:4605–4614
Swanson MM, Fraser G, Daniell TJ, Torrance L, Gregory PI, Taliansky M (2009) Viruses in soils: morphological diversity and abundance in the rhizosphere. Ann Appl Biol 155:51–60
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729
Vos M, Birkett PJ, Birch E, Griffiths RI, Buckling A (2009) Local adaptation of bacteriophages to their bacterial hosts in soil. Science 325:833–833
Whon TW, Kim M-S, Roh SW, Shin N-R, Lee H-W, Bae J-W (2012) Metagenomic characterization of airborne viral DNA diversity in the near-surface atmosphere. J Virol 86:8221–8231
Williamson KE, Radosevich M, Wommack KE (2005) Abundance and diversity of viruses in six Delaware soils. Appl Environ Microbiol 71:3119–3125
Williamson SJ, Allen LZ, Lorenzi HA, Fadrosh DW, Brami D, Thiagarajan M, McCrow JP, Tovchigrechko A, Yooseph S, Venter JC (2012) Metagenomic exploration of viruses throughout the Indian Ocean. PLoS One 7:e42047
Yoshida M, Takaki Y, Eitoku M, Nunoura T, Takai K (2013) Metagenomic analysis of viral communities in (hado) pelagic sediments. PLoS One 8:e57271
Zablocki O, van Zyl L, Adriaenssens EM, Rubagotti E, Tuffin M, Cary SC, Cowan D (2014) High-level diversity of tailed phages, eukaryote-associated viruses, and virophage-like elements in the metaviromes of Antarctic soils. Appl Environ Microbiol 80:6888–6897
Zablocki O, Adriaenssens EM, Cowan D (2016) Diversity and ecology of viruses in hyperarid desert soils. Appl Environ Microbiol 82:770–777
Acknowledgments
This work was supported by the National Science Foundation of China (Grant Nos. 41571248 and 41301265).
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible editor: Zhihong Xu
Rights and permissions
About this article
Cite this article
Han, LL., Yu, DT., Zhang, LM. et al. Unique community structure of viruses in a glacier soil of the Tianshan Mountains, China. J Soils Sediments 17, 852–860 (2017). https://doi.org/10.1007/s11368-016-1583-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11368-016-1583-2