Abstract
Purpose
As artificial islands created by accumulation of sediments and litters, chinampas formed a unique and sustainable agro-ecosystem. However, no investigation on the chinampa microbial communities has been reported. With the goal of revealing the soil bacterial communities in the chinampas and their changes influenced by environmental conditions, soils were sampled for determining their bacterial communities.
Materials and methods
Soil samples were collected from a cultivated and an abandoned chinampa at two horizontal layers and from rhizosphere of Portulaca oleracea L. The bacterial community composition was assayed by 454 high-throughput pyrosequencing. The correlation between environmental parameters and bacterial diversity was analyzed.
Results and discussion
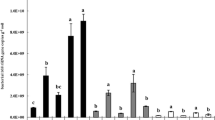
Sequence analysis based on the V1–V3 regions of 16S rRNA gene obtained 140,790 bacterial tags. A total of 22 phyla and 30 candidate divisions were detected in the chinampa soils. The dominant phyla were Proteobacteria, Chloroflexi, Firmicutes, Bacteroidetes, and Acidobacteria. Bacillus, Thiobacillus, Nitrospira, and Planctomyces were the principal genera. Greater bacterial diversity was revealed in the superficial soils than in the deep-layer soils and in bulk soils than in the rhizosphere. The structure of microbial communities in the rhizosphere was remarkably different from that of communities in the bulk soils. Canonical correspondence analysis (CCA) revealed that contents of nitrogen were negatively correlated with Chlorobi, Verrucomicrobia, Gemmatimonadetes, and Acidobacteria but favored Cyanobacteria, Bacteroidetes, and Nitrospirae, whereas total organic carbon and electrical conductivity were positively correlated with Actinobacteria and Chloroflexi.
Conclusions
As the first exhaustive census on the bacterial communities in chinampa soils, our study demonstrates that the chinampas harbor diverse bacteria from soil and sediments and the agricultural activity and rhizosphere effect can shape the microbial communities in this singular agro-ecosystem.




Similar content being viewed by others
References
Alcántara-Onofre S (2005) The floating gardens in Mexico Xochimilco, world heritage risk site. City Time 1:47–57
Altieri MA, Nicholls CI (2013) The adaptation and mitigation potential of traditional agriculture in a changing climate. Climatic Change (Special Issue on Climate Change Mitigation and Adaptation with Local Communities and Indigenous Peoples, edited by McLean KG, Castillo AR, Castellanos E, Lynge A) doi: 10.1007/s10584-013-0909-y
Ansola G, Arroyo P, Sáenz de Miera LE (2014) Characterization of the soil bacterial community structure and composition of natural and constructed wetlands. Sci Total Environ 473–474:63–71
Arndt D, Xia J, Liu Y, Zhou Y, Guo AC, Cruz JA, Sinelnikov I, Budwill K, Nesbø CL, Wishart DS (2012) METAGENassist: a comprehensive web server for comparative metagenomics. Nucleic Acids Res 40:W88–W95
Arroyo P, Sáenz de Miera LE, Ansola G (2015) Influence of environmental variables on the structure and composition of soil bacterial communities in natural and constructed wetlands. Sci Total Environ 506–507:380–390
Bandounas L, Wierckx NJ, de Winde HJ, Ruijssenaars HJ (2011) Isolation and characterization of novel bacterial strains exhibiting ligninolytic potential. BMC Biotechnol 11:94
Basak P, Majumder NS, Nag S, Bhattacharyya A, Roy D, Chakraborty A, SenGupta S, Roy A, Mukherjee A, Pattanayak R, Ghosh A, Chattopadhyay D, Bhattacharyya M (2015) Spatiotemporal analysis of bacterial sediments of Sundarbans using parallel 16S rRNA gene tag sequencing. Microb Ecol 69:500–511
Bellini MI, Gutiérrez L, Tarlera S, Scavino AF (2013) Isolation and functional analysis of denitrifiers in an aquifer with high potential for denitrification. Syst Appl Microbiol 36:505–516
Buée M, De Boer W, Martin F, Van OL, Jurkevitch E (2009) The rhizosphere zoo: an overview of plant-associated communities of microorganisms, including phages, bacteria, archaea, and fungi, and of some of their structuring factors. Plant Soil 321:189–212
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R (2010a) QIIME allows analysis of high–throughput community sequencing data. Nat Methods 7:335–336
Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GI, Knight R (2010b) PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26:266–267
Ceja-Navarro JA, Rivera-Orduña FN, Patiño-Zúñiga L, Vila-Sanjurjo A, Crossa J, Govaerts B, Dendooven L (2010) Phylogenetic and multivariate analyses to determine the effects of different tillage and residue management practices on soil bacterial communities. Appl Environ Microbiol 76:3684–3691
Chan BC, Han XQ, Lui SL, Wong CW, Wang TB, Cheung DW, Cheng SW, Ip M, Han SQ, Yang XS, Jolivalt C, Lau CB, Leung PC, Fung KP (2015) Combating against methicillin-resistant Staphylococcus aureus—two fatty acids from purslane (Portulaca oleracea L.) exhibit synergistic effects with erythromycin. J Pharm Pharmacol 67:107–116
Crossley PL (2004) Sub-irrigation in wetland agriculture. Agri Hum Val 21:191–205
Crouch SR, Malmstdt HV (1967) Mechanistic investigation of molybdenum blue method for determination of phosphate. Anal Chem 39:1084–1089
Doran JW, Elliott ET, Paustian K (1998) Soil microbial activity, nitrogen cycling, and long-term changes in organic carbon pools as related to fallow tillage management. Soil Till Res 49:3–18
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461
Embarcadero-Jiménez S, Yang FL, Freye-Hernández R, Trujillo-Cabrera Y, Rivera Orduña FN, Yuan HL, Wang ET (2014) An improved protocol for extraction of metagenomic DNA from high humus, alkaline and saline soil of chinampa for T-RFLP fingerprinting analysis. Bri Microbiol Res J 4:821–830
Faith DP, Baker AM (2007) Phylogenetic diversity (PD) and biodiversity conservation: some bioinformatics challenges. Evol Bioinform Online 2:121–128
Fierer N, Jackson RB (2006) The diversity and biogeography of soil bacterial communities. Proc Natl Acad Soc USA 103:626–631
Gans J, Wolinsky M, Dunbar J (2005) Computational improvements reveal great bacterial diversity and high metal toxicity in soil. Science 309:1387–1389
Haaijer SC, Van der Welle ME, Schmid MC, Lamers LPM, Jetten MSM, Op den Camp HJ (2006) Evidence for the involvement of betaproteobacterial Thiobacilli in the nitrate-dependent oxidation of iron sulphide minerals. FEMS Microbiol Ecol 58:439–448
Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, Ciulla D, Tabbaa D, Highlander SK, Sodergren E, Methé B, DeSantis TZ, Human Microbiome C, Petrosino JF, Knight R, Birren BW (2011) Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res 21:494–504
Hamady M, Lozupone C, Knight R (2010) Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J 4:17–27
Hammer O, Harper DAT, Ryan PD (2001) PAST: paleontological statistics software package for education and data analysis. Paleontol Electron 4:1–9
Kersters K, De Vos P, Gillis M, Swings J, Vandamme P, Stackebrant E (2006) Introduction to the Proteobacteria. In: Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E (eds) The prokaryotes, vol 5, 3rd edn. Springer, New York, pp 3–37
Krzmarzick MJ, Cray BB, Harding JJ, Oyerinde OO, Leri AC, Myneni SC, Novak PJ (2012) Natural niche for organohalide-respiring Chloroflexi. Appl Environ Microbiol 78:393–401
Kumar A, Prakash A, Johri BN (2011) Bacillus as PGPR in crop ecosystem. In: Maheshwari DK (ed) Bacteria in agrobiology: crop ecosystems. Springer, Berlin, pp 37–59
Lauber CL, Hamady M, Knight R, Fierer N (2009) Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl Environ Microbiol 75:5111–5120
Lienhard P, Terrat S, Chemidlin N, Bouré P, Nowak V, Régnier T, Sayphoummie S, Panyasiri K, Tivet F, Mathieu O, Levêque J, Maron PA, Ranjard L (2013) Pyrosequencing evidences the impact of cropping on soil bacterial and fungal diversity in Laos tropical grassland. Agron Sustain Dev 34:525–533
Ligi T, Oopkaup K, Truu M, Preem J-K, Nõlvak H, Mitsch WJ, Mander Ü, Truu J (2014) Characterization of bacterial communities in soil and sediment of a created riverine wetland complex using high-throughput 16S rRNA amplicon sequencing. Ecol Eng 72:56–66
Merlín-Uribe Y, González-Esquivel CE, Contreras-Hernández A, Zambrano L, Moreno-Casasola P, Astier M (2013) Environmental and socio-economic sustainability of chinampas (raised beds) in Xochimilco, Mexico City. Int J Agr Sustain 11:216–233
Muyzer G, Stams AJ (2008) The ecology and biotechnology of sulphate-reducing bacteria. Nat Rev Microbiol 6:441–454
Nannipieri P, Ascher J, Ceccherini MT, Landi L, Pietramellara G, Renella G (2003) Microbial diversity and soil functions. Eur J Soil Sci 54:655–670
Navarro-Noya YE, Gómez-Acata S, Montoya-Ciriaco N, Rojas-Valdez R, Suárez-Arriaga MC, Valenzuela-Encinas C, Jiménez-Bueno N, Verhulst N, Govaerts B, Dendooven L (2013) Relative impacts of tillage, residue management and crop-rotation on soil bacterial communities in a semi-arid agro-ecosystem. Soil Biom Biochem 65:86–95
Peralta RM, Ahn C, Gillevet PM (2013) Characterization of soil bacterial community structure and physicochemical properties in created and natural wetlands. Sci Total Environ 15:725–732
Price MN, Dehal PS, Arkin AP (2009) FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 26:1641–1650
Qi X, Wang E, Xing M, Zhao W, Chen X (2012) Rhizosphere and non-rhizosphere bacterial community composition of the wild medicinal plant Rumex patientia. World J Microbiol Biotechnol 28:2257–2265
Ramos-Bello R, García-Calderon NE, Ortega-Escobar HM, Krasilnikov P (2011) Artificial chinampas soils of Mexico City: their properties and salinization hazards. Span J Soil Sci 1:71–85
Roesch LFW, Fulthorpe RR, Riva A, Casella G, Hadwin AKM, Kent AD, Daroub SH, Camargo FAO, Farmerie WG, Triplett EW (2007) Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J 1:283–290
Scherf U, Ross DT, Waltham M, Smith LH, Lee JK, Tanabe L, Kohn KW, Reinhold WC, Myers TG, Andrews DT, Scudiero DA, Eisen MB, Sausville EA, Pommier Y, Botstein D, Brown PO, Weinstein JN (2000) A gene expression database for the molecular pharmacology of cancer. Nat Genet 24:236–244
Stroobants A, Degrune F, Olivier C, Muys C, Roisin C, Colinet G, Bodson B, Portetelle D, Vandenbol M (2014) Diversity of bacterial communities in a profile of a winter wheat field: known and unknown members. Microb Ecol 68:822–833
Trujillo-Cabrera Y, Ponce-Mendoza A, Vásquez-Murrieta MS, Rivera-Orduña FN, Wang ET (2013) Diverse cellulolytic bacteria isolated from the high humus, alkaline-saline chinampa soils. Ann Microbiol 63:779–792
Uroz S, Buée M, Murat C, Frey-Klett P, Martin F (2010) Pyrosequencing reveals a contrasted bacterial diversity between oak rhizosphere and surrounding soil. Environ Microbiol Rep 2:281–288
Vásquez-Murrieta MS, Migueles-Garduño I, Franco-Hernández O, Govaerts B, Dendooven L (2006) C and N mineralization and microbial biomass in heavy-metal contaminated soil. Eur J Soil Biol 42:89–98
Verástegui-Valdés MM, Zhang YJ, Rivera-Orduña FN, Cheng HP, Sui XH, Wang ET (2014) Microsymbionts of Phaseolus vulgaris in acid and alkaline soils of Mexico. Syst Appl Microbiol 37:605–612
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267
Wang Y, Sheng H-F, He Y, Wu JY, Jiang Y-X, Tam NF-Y, Zhou H-W (2012) Comparison of the levels of bacterial diversity in freshwater, intertidal wetland, and marine sediments by using millions of illumina tags. Appl Environ Microbiol 78:8264–8271
Whipps JM (2001) Microbial interactions and biocontrol in the rhizosphere. J Exp Bot 52:487–511
Xiang L, Xing D, Wang W, Wang R, Ding Y, Du L (2005) Alkaloids from Portulaca oleracea L. Phytochemistry 66:2595–2601
Xing J, Yang Z, Lv B, Xiang L (2008) Rapid screening for cyclo-dopa and diketopiperazine alkaloids in crude extracts of Portulaca oleracea L. using liquid chromatography/tandem mass spectrometry. Rapid Commun Mass Spectrom 22:1415–1422
Yun J, Ju Y, Deng Y, Zhang H (2014) Bacterial community structure in two permafrost wetlands on the Tibetan Plateau and Sanjiang Plain, China. Microb Ecol 68:360–369
Zhang W, Wu X, Liu G, Chen Y, Zhang G, Dong Z, Yang X, Hu P (2013) Pyrosequencing reveals bacterial diversity in the rhizosphere of three Phragmites australis ecotypes. Geomicrobiol J 30:593–599
Acknowledgments
We acknowledge the assistance of Dr. Olivia Franco Hernández from IPN-UPIBI by her help in the physicochemical analysis of soil. We also thank Dr. M. Soledad Vásquez Murrieta for her help in CCA and Dr. Gerardo Zúñiga from IPN ENCB for his suggestions and review of the manuscript. This research is financially supported by the projects of SIP20140124, authorized by IPN, and 169494 authorized by CONACyT, Mexico. SEJ received a PhD scholarship by the CONACyT (grant number 230841).
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible editor: Jizheng He
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 1581 kb)
Rights and permissions
About this article
Cite this article
Embarcadero-Jiménez, S., Rivera-Orduña, F.N. & Wang, E.T. Bacterial communities estimated by pyrosequencing in the soils of chinampa, a traditional sustainable agro-ecosystem in Mexico. J Soils Sediments 16, 1001–1011 (2016). https://doi.org/10.1007/s11368-015-1277-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11368-015-1277-1