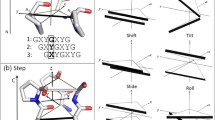
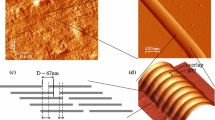
Abstract
Collagen is a unique structural protein that imparts tensile strength to bone, tendons, and numerous other tissues. Like many biological polymers, collagen is continually synthesized and degraded in the extracellular space. While collagen degradation is a normal part of collagen homeostasis, excessive collagenolysis has been implicated in a number of human diseases such as arthritis, cancer, and atherosclerosis. In this work we demonstrate how molecular simulations can be used to study the mechanics of collagen degradation. Dynamical simulations, which model the structural fluctuations that collagen can undergo under physiologic conditions, reveal that portions of collagen are quite flexible—a somewhat counterintuitive finding. Moreover, this flexibility likely facilitates the recognition and cleavage of collagen by proteolytic enzymes. Experiments on collagen-like model compounds are consistent with these observations and demonstrate that new insights into the physical basis of collagenolysis can be obtained from a combination of experiment and computation. More importantly, these results highlight new avenues for the development of potential therapies for disorders that involve abnormal collagen catabolism.










Similar content being viewed by others
References
Voet D, Voet JG (2004) Biochemistry, 3rd edn. Wiley, New York, NY.
Branden C-I, Tooze J (1999) Introduction to protein structure, 2nd edn. Garland, New York, NY.
Lodish HF, Matsudaira PT, Kaiser C, Krieger M (2004) Molecular cell biology, 5th edn. Freeman, New York, NY.
Thomas JM, Williams RJ (2005) Catalysis: principles, progress, prospects. Philos Trans A Math Phys Eng Sci 363:765–791 (discussion 1035–1040).
Hohenester E, Engel J (2002) Domain structure and organisation in extracellular matrix proteins. Matrix Biol 21:115–128.
DeLano WL (2002) The PyMOL Molecular Graphics System. DeLano Scientific, Palo Alto, CA.
Rich A, Crick FH (1961) The molecular structure of collagen. J Mol Biol 3:483–506.
Brodsky B, Ramshaw JA (1997) The collagen triple-helix structure. Matrix Biol 15:545–554.
Gelse K, Poschl E, Aigner T (2003) Collagens—structure, function, and biosynthesis. Adv Drug Deliv Rev 55:1531–1546.
Celentano DC, Frishman WH (1997) Matrix metalloproteinases and coronary artery disease: a novel therapeutic target. J Clin Pharmacol 37:991–1000.
McDonnell S, Morgan M, Lynch C (1999) Role of matrix metalloproteinases in normal and disease processes. Biochem Soc Trans 27:734–740.
Barnes MJ, Farndale RW (1999) Collagens and atherosclerosis. Exp Gerontol 34:513–525.
Fields GB (1991) A model for interstitial collagen catabolism by mammalian collagenases. J Theor Biol 153:585–602.
Lang R, Braun M, Sounni NE, Noel A, Frankenne F, Foidart JM, Bode W, Maskos K (2004) Crystal structure of the catalytic domain of MMP-16/MT3-MMP: characterization of MT-MMP specific features. J Mol Biol 336:213–225.
Stultz CM (2002) Localized unfolding of collagen explains collagenase cleavage near imino-poor sites. J Mol Biol 319:997–1003.
Overall CM (2002) Molecular determinants of metalloproteinase substrate specificity: matrix metalloproteinase substrate binding domains, modules, and exosites. Mol Biotechnol 22:51–86.
Bode W, Maskos K (2003) Structural basis of the matrix metalloproteinases and their physiological inhibitors, the tissue inhibitors of metalloproteinases. Biol Chem 384:863–872.
Humphrey W, Dalke A, Schulten K (1996) VMD: visual molecular dynamics. J Mol Graph 14:33–38, 27–38.
Murphy G, Knauper V (1997) Relating matrix metalloproteinase structure to function: why the “hemopexin” domain? Matrix Biol 15:511–518.
Sacca B, Fiori S, Moroder L (2003) Studies of the local conformational properties of the cell-adhesion domain of collagen type IV in synthetic heterotrimeric peptides. Biochemistry 42:3429–3436.
Hirschfelder J, Eyring H, Topley B (1936) Reactions involving hydrogen molecules and atoms. J Chem Phys 4:170–177.
Alder B, Wainwright T (1959) Studies in molecular dynamics. I. General Method. J Chem Phys 31:459–466.
Rahman A (1964) Correlation in the motion of atoms in liquid argon. Phys Rev 136:A405-A411.
Levitt M, Warshel A (1975) Computer simulation of protein folding. Nature 253:694–698.
McCammon JA, Gelin BR, Karplus M (1977) Dynamics of folded proteins. Nature 267:585–590.
Bernstein FC, Koetzle TF, Williams GJ, Meyer EF, Jr., Brice MD, Rodgers JR, Kennard O, Shimanouchi T, Tasumi M (1977) The protein data bank. A computer-based archival file for macromolecular structures. Eur J Biochem 80:319–324.
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The protein data bank. Nucleic Acids Res 28:235–242.
Brunger AT, Karplus M (1988) Polar hydrogen positions in proteins: empirical energy placement and neutron diffraction comparison. Proteins 4:148–156.
Neria E, Fisher S, Karplus M (1996) Simulation of activation energies in molecular systems. J Chem Phys 105:1902–1921.
MacKerell AD, Jr., Bashford D, Bellot M, Dunbrack RL, Jr., Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, K. K, Lau FTK, Mattos C, S. M, Ngo T, Nguyen DT, Prodhom B, Reiher WE, III, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiórkiewicz-Kuczera J, Yin D, Karplus M (1998) All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B 102:3586–3616.
Huang N, MacKerell AD, Jr. (2004) Atomistic view of base flipping in DNA. Philos Trans A Math Phys Eng Sci 362:1439–1460.
Beveridge DL, DiCapua FM (1989) Free energy via molecular simulation: applications to chemical and biomolecular systems. Annu Rev Biophys Biophys Chem 18:431–492.
McCammon JA, Harvey SC (1987) Dynamics of proteins and nucleic acids. Cambridge University Press, Cambridge.
Straatsma TP, McCammon JA (1991) Theoretical calculations of relative affinities of binding. Methods Enzymol 202:497–511.
Brooks CL III, Karplus M, Pettitt BM (1988) Proteins: a theoretical perspective of dynamics, structure, and thermodynamics. Wiley, New York, NY.
Brooks B, Bruccoleri R, Olafson B, States D, Swaminathan S, Karplus M (1983) Charmm: a program for macromolecular energy, minimization, and molecular dynamics calculations. J Comput Chem 4:187–217.
Brodsky B, Persikov AV (2005) Molecular structure of the collagen triple helix. Adv Protein Chem 70:301–339.
Brooks B III, Bruccoleri R, Olafson B, States D, Swaminathan S, Karplus M (1983) Charmm: a program for macromolecular energy, minimization, and molecular dynamics calculations. J Comput Chem 4:187–217.
Brooks CL III, Karplus M (1983) Deformable stochastic boundaries in molecular dynamics. J Chem Phys 79:6312–6325.
Brooks CL III, Brunger A, Karplus M (1985) Active site dynamics in protein molecules: a stochastic boundary molecular-dynamics approach. Biopolymers 24:843–865.
Kelly SM, Jess TJ, Price NC (2005) How to study proteins by circular dichroism. Biochim Biophys Acta 1751:119–139.
Bhatnagar R, Cough C (1996) Circular dichroism of collagen and related peptides. In: Circular dichroism and the conformational analysis of biomolecules. Plenum, New York.
Venyaminov S, Yang J (1996) Determination of protein secondary structure. In: Circular dichroism and the conformational analysis of biomolecules. Plenum, New York.
Ritort F (2006) Single-molecule experiments in biological physics: methods and applications. J Phys Condens Matter 18:R531–R583.
Greenleaf WJ, Woodside MT, Block SM (2007) High-resolution, single-molecule measurements of biomolecular motion. Annu Rev Biophys Biomol Struct 36:171–190.
Schlick T (2000) Molecular modeling and simulation. An interdisciplinary guide, 1st edn. Springer, New York, NY.
Kramer RZ, Bella J, Mayville P, Brodsky B, Berman HM (1999) Sequence dependent conformational variations of collagen triple-helical structure. Nat Struct Biol 6:454–457.
McQuarrie DA (2000) Statistical mechanics, 1st edn. University Science Books, Sausalito, CA.
Privalov PL, Dragan AI (2007) Microcalorimetry of biological macromolecules. Biophys Chem 126:16–24.
Privalov PL (1982) Stability of proteins. Proteins which do not present a single cooperative system. Adv Protein Chem 35:1–104.
Bahar I, Rader AJ (2005) Coarse-grained normal mode analysis in structural biology. Curr Opin Struct Biol 15:586–592.
Sellers JR, Veigel C (2006) Walking with myosin V. Curr Opin Cell Biol 18:68–73.
Nelson D, Cox M (2005) Lehninger’s principles of biochemistry, 4th edn. Freeman, New York, NY.
Fiori S, Sacca B, Moroder L (2002) Structural properties of a collagenous heterotrimer that mimics the collagenase cleavage site of collagen type I. J Mol Biol 319:1235–1242.
Persikov AV, Ramshaw JA, Brodsky B (2005) Prediction of collagen stability from amino acid sequence. J Biol Chem 280:19343–19349.
Nerenberg P, Salsas-Escat R, Stultz CM (2007) Do collagenases unwind triple-helical collagen prior to peptide bond cleavage? Reinterpreting experimental observations with mathematical models. Protein, Structure, Function, and Bioinformatics. Published online ahead of print: 11 October 2007, DOI 10.1002/prot.21687.
Peterson JT (2004) Matrix metalloproteinase inhibitor development and the remodeling of drug discovery. Heart Fail Rev 9:63–79.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Salsas-Escat, R., Stultz, C.M. The Molecular Mechanics of Collagen Degradation: Implications for Human Disease. Exp Mech 49, 65–77 (2009). https://doi.org/10.1007/s11340-007-9105-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11340-007-9105-1