Abstract
Bacterial leaf blight (BLB), caused by Xanthomonas oryzae pv. oryzae (Xoo), gives rise to devastating crop losses in rice. Disease resistant rice cultivars are the most economical way to combat the disease. The TP309 cultivar is susceptible to infection by Xoo strain PXO99. A transgenic variety, TP309_Xa21, expresses the pattern recognition receptor Xa21, and is resistant. PXO99△raxST, a strain lacking the raxST gene, is able to overcome Xa21-mediated immunity. We used a single extraction solvent to demonstrate comprehensive metabolomics and transcriptomics profiling under sample limited conditions, and analyze the molecular responses of two rice lines challenged with either PXO99 or PXO99△raxST. LC–TOF raw data file filtering resulted in better within group reproducibility of replicate samples for statistical analyses. Accurate mass match compound identification with molecular formula generation (MFG) ranking of 355 masses was achieved with the METLIN database. GC–TOF analysis yielded an additional 441 compounds after BinBase database processing, of which 154 were structurally identified by retention index/MS library matching. Multivariate statistics revealed that the susceptible and resistant genotypes possess distinct profiles. Although few mRNA and metabolite differences were detected in PXO99 challenged TP309 compared to mock, many differential changes occurred in the Xa21-mediated response to PXO99 and PXO99△raxST. Acetophenone, xanthophylls, fatty acids, alkaloids, glutathione, carbohydrate and lipid biosynthetic pathways were affected. Significant transcriptional induction of several pathogenesis related genes in Xa21 challenged strains, as well as differential changes to GAD, PAL, ICL1 and Glutathione-S-transferase transcripts indicated limited correlation with metabolite changes under single time point global profiling conditions.
Similar content being viewed by others
1 Introduction
There’s a growing interest in studies that attempt to link gene expression with metabolite profiling (Brosche et al. 2005; Kaplan et al. 2007; Jumtee et al. 2008; Vijayendran et al. 2008). Many plant functional genomics studies have combined the generation of transgenic and mutant plants with the parallel analysis of mRNA expression, protein levels and plant histology studies (Fiehn et al. 2000; Finkelstein et al. 2002; Oliver et al. 2002; Henikoff and Comai 2003; Farrokhi et al. 2006; Steinberg et al. 2008). Extensive literature also exists for metabolic perturbation of host/plant interactions through parallel analysis of metabolomics experiments and microarray analysis (Urbanczyk-Wochniak et al. 2003; Dharmadi and Gonzalez 2004; Hirai et al. 2004; Verhoeckx et al. 2004; Matsuzaki et al. 2005; Frey et al. 2007; Cho et al. 2008; Hoefgen and Nikiforova 2008; Nam et al. 2009; Tan et al. 2009; Yang et al. 2009). Thus, workflows to integrate metabolic and genomic analysis will contribute to a comprehensive biological understanding for observed phenotypic differences between organisms that gene expression studies alone would otherwise be unable to achieve.
Rice provides food for over half the world’s human population, and the availability of its genome sequence (2005) is important for research and development of disease resistant cultivars. Bacterial leaf blight (BLB) is responsible for major rice production losses in irrigated and rain-fed lowland ecosystems. The use of resistant rice varieties continues to be the most economical and effective way to combat this disease. Cell surface pattern recognition receptors (PRRs) are key components of the innate immune response in animals and plants. The rice PRR, XA21 (Song et al. 1995), recognizes the pathogen associated molecular pattern (PAMP), Ax21 (Activator of XA21-mediated immunity) that is highly conserved in all tested species of the genus Xanthomonas as well as in Xylella species (Lee et al. 2006; Lee et al. 2009). Recently, mechanisms for XA21-mediated resistance have been elucidated (Park et al. 2008).
We used metabolomics and transcriptomics profiling workflows for discovery analysis to contrast the responses of two rice varieties to Xoo: (1) TP309, the parent rice genotype, which is susceptible to infection by the Philippine bacterial Xoo strain PXO99, expressing Ax21 and (2) TP309_Xa21, a transgenic variety of rice harboring the Xa21 PRR, which is resistant to Xoo strain PXO99. Production of Ax21 requires the presence of eight rax (required for activation of Xa21-mediated immunity) gene family members (Shen et al. 2002; Burdman et al. 2004; da Silva et al. 2004), including raxST. Strains carrying mutations in any of the known rax genes are no longer recognized by XA21-containing rice lines resulting in lesion development on XA21-lines. For example, PXO99 strains carrying a knockout in the raxST gene (PXO99△raxST) cause long lesions on TP309_Xa21 lines. In contrast, TP309 is susceptible to both PXO99 and PXO99△raxST strains.
We developed a robust, all-in-one rice leaf sample extraction method that uses a single organic solvent mixture of water/acetonitrile/isopropanol for LC/MS, GC/MS and microarray analyses. Importantly, contaminating waxes that can potentially foul the MS source were not extracted. Our MS data analysis workflow included post-processing data filtering to yield higher quality results that was required to shed further insights into metabolite and transcript perturbations of this plant/parasite interaction.
2 Materials and methods
2.1 Study design
Seven separate groups (or conditions) of rice were examined (Table 1). For the TP309 rice line, three conditions were tested: (1) PXO99 inoculated, (2) mock-treated, (3) non-treated (NT). For the transgenic TP309_Xa21 rice [TP309-106-17-3-37 (Song et al. 1995)], four conditions were studied to compare the response of TP309_Xa21 rice to bacterial inoculation to which it is resistant or susceptible: (4) PXO99 inoculated, (5) mock- treated (6) non-treated and (7) PXO99△raxST inoculated. Biological replicates (n = 6) for each of the experimental conditions was evaluated, except for the non-treated groups where n = 9. PXO99, carrying Ax21 activity and PXO99△raxST strain lacking Ax21 activity were inoculated onto leaves of the rice lines TP309 and TP309_XA21 and analyzed for their metabolomic profiles. Appropriate mock-treated controls were also included. PXO99 and PXO99△raxST were grown for 72 h at 30°C on peptone sucrose agar. Six-week-old rice plants (TP309 and TP309_Xa21) were cut approximately 4 cm from the tip of fully expanded leaves with scissors dipped in PXO99 or PXO99△raxST bacterial suspensions at either 1 × 109 cells per ml (Kauffman et al. 1973) or just peptone sucrose agar (mock treatment). After inoculation, plants were maintained in a growth chamber and allowed to grow for 4 days.
2.2 Rice sample processing and metabolite extraction
The samples were processed according to Weckwerth et al. (2004) with the following changes. Rice leaves were collected under controlled conditions, flash frozen and stored at −80°C. The cutting of reproducible amounts of frozen rice leaf samples for metabolomics and gene expression analysis is critically important. First because the solvent volume and solvent ratios are optimized for samples in this weight range and second because similar reproducible measurements for all groups of samples allows inter-group comparisons of data. 18 ± 2.2 mg segments of rice leaves were cut and weighed. The rice leaves were placed in liquid nitrogen cooled, 2 ml Eppendorf tubes, each containing a 5 mm stainless steel ball bearing from Retsch (Newtown, PA). The tubes were transferred to MM301 Retsch mill adapter racks that had also been pre-cooled in liquid nitrogen. Rice leaves were ground in the homogenizer for 30 s at 25 Hz. Metabolites were extracted from membrane and cell wall components in 1 ml of a single phase, 2:3:3 v/v/v solvent mixture of water/acetonitrile/isopropanol, that had been pre-cooled at −20°C in a Digital One RTE-7 refrigerated circulating bath (Thermo Electron Corp., Newington, NH). This solvent mixture minimizes the extraction of waxes that are detrimental to the analytical instrumentation and enables analysis of metabolites by both LC–TOF and GC–TOF instruments. After the samples were incubated at −20°C in the refrigerated circulating bath for 20 min, the tubes were centrifuged to pellet mRNA and protein and were stored at −80°C for subsequent mRNA processing. The liquid phase supernatant extracts were divided equally and transferred under nitrogen gas to autosampler vials (Agilent, Palo Alto, California, USA) with Teflon/silicon septa, for LC–TOF or GC–TOF analyses.
2.3 Total RNA extraction, RNA quality analysis, amplification and labeling
Total RNA was extracted from the cell pellets using the Agilent Plant RNA Isolation Mini Kit (Agilent Technologies, Santa Clara, CA). This phenol-free spin column technique results in RNA with substantially reduced genomic DNA (gDNA) contamination without the need for DNase I treatment. Briefly, the pellet was extracted in 4 M guanidine hydrochloride, 25 mM Tris–HCl and β-mercaptoethanol solution, spun through a pre-filtration column and RNA was retained on a mini isolation column. The RNA was washed, dried and eluted off the column in 10–50 μl of nuclease-free water.
The NanoDrop® ND-1000 Spectrophotometer was first used to determine the concentration of purified RNA. The Agilent 2100 Bioanalyzer was used with the RNA 6000 Nano LabChip® Kit (Agilent Technologies, Santa Clara, CA), which uses fluorescent dyes to bind nucleic acids for tracking the electrophoretic separation of total RNA. One micro liter of each sample (25–500 ng/μl) was loaded per well to assess ribosomal RNA quality by an accurate reading of 5S, 18S and 28S peaks, along with any degradation. The Low RNA Input Linear Amplification Kit PLUS, Two-Color kit (Agilent Technologies, Santa Clara, CA) generated fluorescent complimentary RNA (cRNA) for use in hybridizing with oligonucleotide microarrays. Briefly, for each sample, 500 ng of extracted Total RNA was first mixed with two spike mixes from the Agilent Two-Color RNA Spike-In Kit (Agilent Technologies, Santa Clara, CA) according to manufacturer instructions. Each mix contains 10 in vitro synthesized, polyadenylated transcripts in predetermined ratios that in the subsequent hybridization process specifically binds to control probes on a microarray. The results are intended to provide expected versus observed log ratios for monitoring microarray hybridization linearity, sensitivity, and accuracy. After adding the two spike mixes, a primer containing poly dT and a T7 polymerase promoter was added to the mixture, and annealed to the poly A + RNA. The first and second strands of cDNA were synthesized by adding reverse transcriptase to the mix. Next, T7 RNA polymerase was added to synthesize cRNA from the double-stranded cDNA, and simultaneously incorporated cyanine 3- or cyanine 5-labeled CTP (Qiagen N.V., Venlo, Netherlands). RNeasy mini spin columns were used to purify the amplified cRNA samples. Fluorescently labeled cRNAs were analyzed on a ND-1000 Spectrometer (NanoDropTechnologies, Rockland, DE) for determining yield and specific activity. The yield for the rice RNA was in the range expected for plant products, with an average specific activity of cyanine 3- and cyanine 5-labeled cRNA targets of 14.6 (±3.3) pmol dye/μg cRNA.
2.4 Microarray analysis
The Agilent Gene Expression Hybridization Kit (Agilent Technologies, Santa Clara, CA) was used for hybridizing fluorescently-labeled cRNA to the Agilent 22 K Rice oligo microarrays according to the manufacturer instructions. This 22,575 probe microarray contains 60-mer probes, with broad gene coverage, representing an estimated 60% of the rice genome (japonica race). The probes were designed from the extensive cDNA library for rice, developed by Dr. Shoshi Kikuchi of Japan’s National Institute of Agrobiological Sciences (NIAS). Annotation for this microarray is presented on the NIAS website at (http://cdna01.dna.affrc.go.jp/cDNA/). The 1 × 3′′ glass slides were washed, dried and scanned using an Agilent laser scanner according to manufacturer instructions. Agilent Feature Extraction software was used for automated processing of microarray image files that included dye normalization using LOWESS, filtering of poor, flagged pixels and determination of feature intensities and ratios. The output files were imported into GeneSpring-GX data analysis software, where the data was normalized to the 50th percentile across the different slides. The replicate dye-swapped samples were grouped into one experiment and analyzed for statistically significant differential genes. To find differentially expressed genes between two different conditions, the gene lists were further filtered using Volcano plots, which permitted the visualization of the relationship between fold-change and statistical significance (which takes both fold change and P-value into consideration).
2.5 GC–TOF mass spectrometry
A mixture of internal retention index (RI) markers was prepared using fatty acid methyl esters of C8, C9, C10, C12, C14, C16, C18, C20, C22, C24, C26, C28 and C30 linear chain length, dissolved in chloroform at a concentration of 0.8 mg/ml (C8–C16) and 0.4 mg/ml (C18–C30). 1 μl of this RI mixture was added to the dried extracts. 10 μl of a solution of 20 mg/ml of 98% pure methoxyamine hydrochloride (Sigma, St.Louis, MO) in pyridine (silylation grade, Pierce, Rockford, IL) was added and shaken at 30°C for 90 min to protect aldehyde and ketone groups. 90 μl of MSTFA.1%TMCS (1 ml bottles, Pierce, Rockford, IL) was added for trimethylsilylation of acidic protons and shaken at 37°C for 30 min. The reaction mixture was transferred to a 2 ml clear glass autosampler vial with microinsert (Agilent, Santa Clara, CA). A Gerstel automatic liner exhange system was used in conjunction with a Gerstel CIS cold injection system (Gerstel, Muehlheim, Germany). An Agilent 6890 gas chromatograph (Santa Clara, CA) was controlled by the Leco ChromaTOF software versus 2.32 (St. Joseph, MI). A 30 m long, 0.25 mm i.d. rtx5Sil-MS column with 0.25 μm 5% diphenyl/95% dimethyl polysiloxane film and additional 10 m integrated guard column was used (Restek, Bellefonte, PA). Result *.txt files were exported to a data server with absolute spectra intensities and further processed by a filtering algorithm implemented in the metabolomics BinBase database (Fiehn et al. 2005). Quantification was reported as peak height using the unique ion as default, unless a different quantification ion was manually set in the BinBase administration software Bellerophon. A quantification report table was produced for all database entries that were positively detected in more than 50% of the samples of a study design class as defined in the SetupX database (Scholz 2007). A subsequent post-processing module was employed to automatically replace missing values from the *.cdf files using libraries obtained from the open access mzmine software. Replaced values were labeled as ‘low confidence’ by color coding.
Metabolites were unambiguously assigned by the BinBase identifier numbers, using retention index and mass spectrum as the two most important identification criteria. Additional confidence criteria were given by mass spectral metadata, using the combination of unique ions, apex ions, peak purity and signal/noise ratios as given in data preprocessing. All database entries in BinBase were matched against the Fiehn mass spectral library of 1,200 authentic metabolite spectra and the NIST05 commercial library (Kind et al. 2009). BinBase entries were named manually by both matching mass spectra and retention index and PubChem numbers and KEGG identifiers were added. In addition, all reported compounds (identified and unknown metabolites) are reported by the quantification ion and the full mass spectrum encoded as string. All raw and processed data are downloadable at http://fiehnlab.ucdavis.edu:8080/m1/main_public.jsp.
2.6 LC–TOF mass spectrometry
An Agilent 1100 LC equipped with a ZORBAX SB-Aq column 2.1 × 150 mm, 3.5 μm was used to separate the rice extracts. The LC parameters were as follows: autosampler temperature, 4°C; injection volume, 2 μl; column temperature, 20°C; and flow rate of 0.3 mL/min. The LC solvent and time table were: a 2–98% linear gradient of water/acetonitrile employed over 46 min followed by a solvent hold until 54.9 min, at which time data collection was stopped. 0.1% formic acid was used as a mobile phase modifier. Mobile phase: A = 0.1% formic acid in water, B = 0.1% formic acid in acetonitrile. Gradient: 2% B at 0 min., 98% B at 46 min., 98% B at 54.9 min., 2% B at 55 min. MS stop time: 54.9 min. LC stop time: 55 min.
Mass spectrometric analysis was performed on an Agilent 6210 Time-of-Flight LC/MS equipped with an electrospray (ESI) ion source. Nitrogen was used as the drying gas at 35 psi. The ESI source used a separate nebulizer for the continuous, low-level (10 μl/min) introduction of reference mass compounds: 121.050873, 922.009798 (positive ion mode) and 119.036320, 966.000725 (negative ion mode). ESI spray voltage was 4,000 V in positive mode and 3,500 V in negative ion mode and fragmentor at 170 V. Nitrogen was used as sheath gas at 30 psi. The accurate-mass LC–TOF chromatograms were acquired and deconvoluted into individual chemical peaks. The molecular features were identified using the Molecular Feature Extraction (MFE) algorithm in MassHunter™ software (Agilent Technologies, Santa Clara, CA, USA). The retention time/mass pairs generated by the MassHunter™ software were then exported for subsequent statistical analysis.
2.7 Statistics
GeneSpring-MS 1.2 statistical analysis software was used for retention time and mass abundance adjustments. The multiple data sets were normalized to correct data for retention time and response drift, and aligned before they were compared. The workflow followed the following steps: (1) Alignment and normalization of features across all samples. (2) Data Transformation: measurements less than 0.01 were set to 0.01; (3) Normalization: “Per Mass” normalized to the median, (4) Filtering: the six independent biological replicate samples per group were analyzed as one group after the ion intensity for each molecular ion was averaged across the six biological replicates. The relative frequency of detection (number of samples a particular feature is found in one or more groups) was varied and compared. The highest quality features analyzed were those that were only present in all six replicates, in at least one or more groups.
The features were then analyzed by Volcano plots to reveal both fold changes and statistically significant (P < 0.05) differential abundance profiles. The resulting mass lists were then queried against the METLIN Personal Metabolite database to find matches based on neutral mass, using a 5 ppm window. This database currently contains over 23,000 compounds, and includes links to KEGG identifiers, CAS numbers, HMDB and LipidMaps. Because retention time information was not used as an orthogonal criterion for identification, a Molecular Formula Generator (MFG) algorithm was used to calculate an independent best fit empirical formula for each feature using the mass spectral metadata: monoisotopic mass, isotope ratio and abundance information. The MFG score helped in determining whether the calculated empirical formula agreed with the formula for the compound in the database match. Isomers and isobaric compound remained unresolved. Statistical evaluation was performed using univariate analyses, including the Student t-test for independent pairs of groups, and one way ANOVA for multiple groups. The effect of the treatment (example, mock-treated and PXO99-infected) and rice lines (TP309 and TP309_Xa21) were compared. Because PXO99△raxST was an unpaired condition that was only tested against the TP309_Xa21 line, it was independently compared to mock-treatment in the TP309_Xa21 line using a t-test. Values of P < 0.05 were considered statistically significant and the Benjamini and Hochberg (Benjamini and Hochberg 1995) False Discovery Rate of 5% was used for multiple testing correction. We then investigated which of the mean(s) in the ANOVA were different using the Tukey (Tukey 1949) multiple comparison test. Multivariate statistical analysis was performed by unsupervised principal component analysis (PCA) with mean centering and scaling to visually demonstrate the variance of the metabolic phenotypes from the groups based on the discriminating features from the ANOVA analysis. Hierarchical cluster analysis of metabolites was performed to reveal associations between replicate biological samples within a group based on the similarity of their mass abundance profiles.
For GC–TOF data, statistical evaluations were performed by Statistica DataMiner versus 7.0 (StatSoft, Tulsa, OK). For univariate statistics, data were log(10)-transformed to reduce weight of outliers and to render the data sets more normal-distributed. Two-tailed Student t-tests were performed with significance thresholds of P < 0.05. Ratios were calculated from antiloged class averages. For PCA and Partial Least Square (PLS) analyses, variables were scaled to unit standard deviations and components were calculated with the Non Linear Iterative PLS algorithm using 50 iterations and a 0.0001 convergence criterion with a 7-fold cross-validation for supervised PLS. PCA was carried out on unit variance scaled data in unsupervised mode for dimension reduction purposes.
3 Results and discussion
The susceptible cultivar TP309 was compared to the TP309_Xa21 transgenic variety in the absence of infection to see whether the transgenic event itself triggered metabolic differences leading to resistance. We further investigated metabolomic data to study if the TP309_Xa21 plants show specific metabolic responses after infection that might contribute to the resistance. As a positive control we used PXO99△raxST bacteria that are virulent on TP309_Xa21 plants in order to discriminate metabolic differences due to the transgenic host and perturbations that are caused by the infection.
3.1 The number of detected peaks or features increases linearly with class frequency threshold reduction
Often, biological replicates of individual plants grown under otherwise identical conditions nevertheless show biological variability that is larger than the pure technical data acquisition variability. We therefore calculated the relationship between the “class frequency threshold” (% relative frequency of features detected across replicates in at least one group), in reverse phase LC–TOF and liquid injection GC–TOF analyses, and the number of reported features. Table 2 shows the number of features roughly increases linearly with decreasing of the class frequency threshold. The reduction in feature numbers with higher threshold stringency certainly improves the confidence in subsequent statistical investigations by limiting the dimensionality problem and by generally de-noising the data.
In total, 514 unfiltered features were detected in LC–TOF runs in positive ion mode and 474 in negative ion mode. Upon filtering, the features were reduced to 355 (50% threshold) or 186 (100% threshold). Secondly, it was apparent that after comprehensive peak picking and constraining result files, a similar total number of peaks for GC were calculated for GC–TOF files, suggesting the complementary nature of these technologies. Furthermore, a similar number of peaks were obtained from negative ionization and positive ionization LC–TOF analyses, indicating that both modes are necessary for discovery metabolomics. In principle, it should be possible to combine negative/positive LC–TOF data files to a non-redundant list of mass features using the retention time/accurate mass information concomitant with lists of ion adducts in both modes. Unfortunately, the software did not support this strategy at the time of analysis. GC–TOF data can be classified to identified and structurally annotated metabolites (via retention index and mass spectral matching to reference libraries), in comparison to structurally uncharacterized unknown metabolites. In a similar manner to the total number of metabolic signals, the number of identified metabolites linearly increased from 104 to 154 non-redundant metabolites with decreasing of relative frequency threshold levels. This finding confirmed that setting the positive relative frequency threshold at 50% was reasonable for comprehensive metabolomics. The fact that only around one-third of all peaks were structurally assigned is typical for unbiased metabolomics screening, suggesting that functional annotation of plant genes would greatly benefit from improved algorithms and libraries used for compound identifications.
3.2 Rice cultivar TP309 and its Xa21 transgenic variant differ in metabolic phenotypes
In order to determine whether inherent differences exist between the two untreated (NT) rice genotypes even before bacterial challenges, we examined their metabolic phenotypes by classic dimension reduction using unsupervised Principal Component Analysis (PCA, Supplement 1). The NT rice line TP309_ XA21 already exerts subtle differences in metabolite expression compared to its genetic background which were apparent using the first three vectors for negative ionization LC–TOF data (60.7% of explained total variance of 80 variables, Supplement S1A) as well as for GC–TOF data (48% of explained total variance of 441 variables, Supplement S1B). Using ANOVA, the measured metabolite abundances for a range of sugar alcohols, some TCA intermediates and miscellaneous compounds were differential between the two genotypes. This was interpreted as a pleiotropic effect of the primary genetic alteration but was not found in pathways that were later determined to be involved in resistance to bacterial infection. As part of our exploratory phase we performed Hierarchical Cluster Analysis (HCA) by grouping the samples into clusters based on the similarity of their metabolite abundance profiles. Figure 1 presents the resulting heat map for all detected metabolites in all samples by LC–TOF under positive electrospray mode. The heat map reveals good clustering of sample replicates and the correlation coefficients emphasize the separation of the different groups, particularly between the two rice genotypes. Interestingly, the samples for the Xa21_genotype infected with PXO99△raxST were divided between TP309 and Xa21 genotypes infected with PX099, indicating a distinctive metabolite profile.
Hierarchical cluster analysis of LC–TOF (+) data of six biological replicates for the seven rice genotype/infection classes. The mass trees (dendrograms) reveal the relationships between the different conditions based on their abundance levels with each rectangle or cell representing an accurate mass, colored by its abundance intensities on a normalized scale from 0 (low) to 5 (high). Masses with similar abundance patterns are placed closer to each other. The mass tree branches at the top of the figure are colored by each condition, and shows the correlations for the main clades or branches. The bottom of the figure shows that the tree branches for each group of conditions are separated based on rice genotype. The * denotes four out of six Xa21-PXO99△raxST samples clustering with the TP309 genotype
3.3 Unsupervised and supervised classification tools clearly discriminate TP309_Xa21 challenged bacterial strains
The most interesting question in this study was whether PX099 challenge induces different metabolic responses in susceptible TP309 versus the resistant TP309_Xa21 genotype, and secondly, if TP309_Xa21 samples respond differently to PXO99△raxST versus PX099.
Although GC–TOF results showed that untreated (NT) TP309 and TP309_Xa21 metabolic phenotypes exerted a number of differences at t-test significance levels of P < 0.05, for example in sugar alcohols, trehalose, raffinose, pipecolic acid, fumaric acid and maleic acid (Supplement S2), none of these compounds were found to be relevant when exploring metabolic responses in the TP309_Xa21 towards PX099 (resistant) and PXO99△raxST (susceptible) bacterial challenges, for which clear metabolic phenotypes were found using both unsupervised and supervised multivariate statistics.
One-way ANOVA with Tukey post-hoc testing on LC–TOF data resulted in many statistically significant masses between the three different conditions in the TP309_Xa21 cultivar (Supplement S3). A total of 44 compounds (Supplement S3A) were significantly different between all pair wise comparisons by LC TOF–MS (+), and 39 compounds (Supplement S3B) by LC TOF–MS (−). The results are graphically divided into cells showing the number of statistically significant (or not significantly different) masses. The largest difference was in positive ion mode between mock and PXO99 challenge, showing 27 significantly different compounds. The ANOVA results for the metabolic phenotypes were then visualized by performing PCA that revealed clear discrimination between the three groups of samples (Fig. 2a).
Supervised multivariate statistics to differentiate bacterial challenge phenotypes for TP309_Xa21 transgenic rice plants. Left panel: LC–TOF (positive ionization) with one-way ANOVA analysis of 212 masses present at a 50% class frequency threshold in three TP309_Xa21 classes. Subsequent dimension reduction and PCA visualization of 44 statistically different (ANOVA) masses (see Supplement S3A). Right panel: GC–TOF analysis of 441 metabolites present at a 50% class frequency threshold. Non Linear Iterative PLS supervised discrimination of TP309_Xa21 classes
For GC–TOF analysis, where we benefited from the BinBase database of known, structurally identified compounds, supervised partial least squares classification was used to distinguish metabolic responses between bacterial challenged and the corresponding mock-treated controls. In both cases, distinct metabolic phenotypes were clearly discriminated (Fig. 2b), showing that responses in TP309_Xa21 were indeed different depending on the strain of bacteria used to infect the plant. Interestingly, the results for both LC–TOF and GC–TOF platforms more of the variance was explained by the general difference of mock-treated and PXO99 challenged plants (vector 1) than between PXO99 versus PXO99△raxST inoculations, discriminated by vector 2. This finding indicates that Xoo infection results in metabolic adaptations even if the strain used does not cause long lesions, and is consistent with observations that Xoo can grow to fairly high levels in the early stages of infection even in resistant Xa21 cultivars (Song et al. 1995; Lee et al. 2009).
Next, detailed pair wise comparisons were performed between the different groups that included fold change and significance analysis based on unpaired t-test. The results are summarized in Table 3. The number of significant metabolic differences between PXO99 and mock-challenged TP309 cultivar was clearly lower than observed for TP309_Xa21. This observation was especially evident for those significant differences that had a larger than 2-fold differences. In GC–TOF and LC–TOF (−) data sets, but not for LC–TOF (+) data, PXO99△raxST-inoculated plants showed a higher number of significantly different metabolites than for PXO99-versus mock-challenged controls. This result was not dependent on the relative frequency of detection thresholds (50, 75 or 100%). Similarly, metabolic differences were higher in LC–TOF (+) than in negative ion mode analyses for the infected TP309 plants. Taken together, the LC–TOF data suggests that compound classes detected in the negative ion mode, such as the organic acids, do not appreciably change in response to PXO99. In addition, PXO99△raxST results imply differential abundance levels for different classes of metabolites compared to the PXO99 strain.
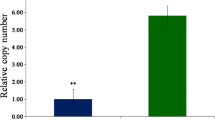
The results of METLIN personal metabolite database matching are summarized in Supplement S4. For each of these compounds, the majority of MFG scores were high confident matches, typically greater than 75%, indicating a good correlation between the MFG derived formula (based on isotopic mass, abundance and spacing) and the METLIN database formula. The result provided the extent of metabolite coverage that currently can be annotated in rice samples using exact mass LC–TOF analysis. Because isomeric and isobaric compounds cannot be differentiated by database query alone, only one representative compound was annotated. Several compounds were significantly elevated in the TP309_Xa21 rice line, after challenge with PXO99 or PXO99△raxST−. These included Rutin, several pigments, fatty acids and lipids. For example, levels of m/z 174.1112 (Supplement S4), matching Arginine, was highly increased (14.6 fold) in Xa21-PXO99 relative to mock-treated rice leaves. Arginine has an important role in plant metabolism via the urea cycle, for example for polyamine biosynthesis (Bassie et al. 2003). Free arginine is derived from endogenous synthesis and in the turnover of proteins. Although synthesis of arginine from citrulline can occur in many cell types (Eagle 1959; Eagle et al. 1959; Jackson et al. 1986), in cases of catabolic stress, such as infection, the levels of endogenous synthesis may not be sufficient to meet metabolic demands. Arginine homeostasis is accomplished by modulation of arginine catabolism rather than synthesis (Morris 2007). Thus one hypothesis to explain this observation is an increased free pool of arginine, generated due to catabolism of pathogen-derived proteins. Ornithine, citrulline, tyrosine, phenylalanine, lysine (determined by GC–TOF) and arginine (determined by LC–MS) all showed increased levels in the (resistant) PXO99-challenged Xa21 genotype compared to mock-treated controls, but not in other (infection-susceptible) comparisons (Fig. 3; Supplement S2). GC–TOF analysis of proline levels were unaffected, which is different to other biotic stress studies, e.g. rice infection by the Tungro virus (Mohanty and Sridhar 1982). The Increases in these metabolite pools suggests that some metabolic pathways may be directly involved in the XA21-mediated defense against pathogen infection.
Box-whisker plots of GC–TOF data of metabolic differences in transgenic TP309_Xa21 rice leaves infected with PXO99 (resistant) or PXO99_ΔraxST (susceptible), compared to mock-treated and to susceptible PXO99-infected TP309 plants. Median levels with whiskers of 1.96× S.E. are given, n = 9 for infected plants and n = 6 for mock controls. Differentially expressed metabolites are listed as Supplement data in S2 for GC–TOF data
3.4 Important precursors for alkaloid metabolism are significantly higher in the resistant TP309_Xa21 genotype challenged with the PXO99 strain
Phenylalanine and tyrosine had been found before to be inducible in response to biotic stress, for example by fungal attacks on plant leaves (Khan et al. 2003). Both compounds are precursors for alkaloid biosynthesis or, alternatively, for the production of phenylpropanoids which are furthered into lignins. Therefore, up-regulation of phenylalanine and tyrosine levels may help the resistant TP309_Xa21 transgenic rice to defend against bacterial infection by alkaloids and partial cell wall lignifications. The induction of phenylalanine- and phenylalanine/tyrosine ammonia lyases (PAL and PTAL, Os04g0518400 and Os02g0626100) is a hallmark of plant responses to bacterial infection and frequently used as indicator or even elicitor of cellular response mechanisms in rice (Bera and Purkayastha 1999; Vidhyasekaran et al. 2001; Jayaraj et al. 2004). Ornithine, citrulline and arginine are all member of the urea cycle that salvage ammonia, which is released by the phenylalanine/tyrosine ammonia lyase reaction, but are also precursors in alkaloid biosynthesis via putrescine or cadaverine intermediates. Hence, it can be concluded that several important precursors for alkaloid metabolism were found to be significantly up-regulated in the resistant PXO99-challenged TP309_Xa21 genotype in comparison to all other test cases including the PXO99△raxST-challenged TP309_Xa21 genotype. A range of further metabolites were significantly differentially regulated when comparing the PXO99△raxST-challenged TP309_Xa21 to both the PXO99-challenge and the mock-treated TP309_Xa21 plants. Some metabolites such as glutamine (Fig. 3) and phenol, ala-ala, 5-hydroxynorvaline and hydroxylamine (Supplement S2) were more than 2-fold higher in the PXO99-challenged Xa21 plants than in the susceptible PXO99△raxST challenge. Concomitantly, in the PXO99△raxST treatment, a range of metabolic differences were apparent that did not occur in any of the other treatments. Figure 3 details a down-regulation of glutamate levels that was found concurrent with up-regulation in two glutamate products, oxoproline and N-acetylglutamate. Oxoproline is formed from glutamyl-cysteine using the cyclizing (5-l-Glutamyl)-l-amino-acid 5-glutamyltransferase (EC 2.3.2.4), yielding cysteine and the cyclized amino acid product. Interestingly, a second cyclized product was observed, butyrolactam, for which yet no enzyme has been identified in rice but which might be produced from GABA (4-aminobutyric acid) using the same enzyme but the hypothetical GABA-cysteine as substrate. Indeed, cysteine was found more than 2-fold up-regulated in the PXO99△raxST treated Xa21 plants (susceptible) compared to both mock-treated and PXO99-treated plants, albeit less significant (P = 0.1). This finding supports the assumed cyclization reaction via EC 2.3.2.4 with cysteine as by-product. The intermediate Glu-Cys is also an important precursor in the biosynthesis of glutathione (Glu-Cys-Gly) which requires glycine as further substrate, and indeed, 3-fold higher levels of glycine were found at P < 0.05 in the PXO99-challenged Xa21 plants (resistant) than in the PXO99△raxST treated or the mock-treated Xa21 plants (Supplement S2). Taken together, these data indicate that the resistance phenotype in transgenic Xa21 plants upon PXO99-challenge is manifested in up-regulation of precursors for alkaloid biosynthesis as well a potential higher flux into glutathione biosynthesis than into the alternative reaction towards cyclization into oxoproline.
3.5 The virulence signal acetophenone is absent in TP309_Xa21-infected rice plants but abundant in mock-treated controls
Upon bacterial challenges, a range of further compounds were found to be differentially regulated in TP309_Xa21 plants (Supplement S2) which do not seem to be directly involved in the resistance phenotype itself. For example, the phytosterols stigmasterol and beta-sitosterol were found at 30% lower levels in PXO99 challenged Xa21 plants compared to PXO99△raxST challenged Xa21 plants or the corresponding mock-treated Xa21 plants, but the same difference was found in susceptible PXO99-challenged TP309 plants, lacking Xa21. In the transgenic Xa21-plants, a 2-fold reduction in the cell wall component sinapinic acid (but not ferulic acid) was found upon treatment with either PXO99△raxST or PXO99, compared to the mock control.
Most strikingly, however, was that acetophenone and its reduction product 2-phenylpropanol were both completely absent in Xa21 plants challenged with either PXO99 or PXO99△raxST (Fig. 4). In contrast, these compounds were detected in TP309 and TP309_ PXO99-treated plants as well as Xa21 mock-treated plants at abundant levels, as well as in TP309 and TP309_Xa21 untreated plant (NT, data not shown). Acetophenone derivatives had previously been implicated as signaling molecules in bacterial virulence pathways and gene expression modulators in Solanaceae species but not in monocotyledonous species (Spencer 1991). Therefore, the complete lack of acetophenone in TP309_Xa21 rice plants challenged with either PX099 strain suggests that Xa21 responds to both Xoo strains differently than it responds to mock treatment. This result indicates the need for additional investigation into the mode of action and biochemical pathways triggered by PRRs, and into acetophenone-related response pathways.
Box-whisker plots of GC–TOF data of metabolic differences in infected transgenic TP309_Xa21 rice leaves, compared to mock-treated and to PXO99-infected TP309 plants. Median levels with whiskers of 1.96× S.E. are given, n = 9 for infected plants and n = 6 for mock controls. Differentially expressed metabolites are listed as Supplement data in S2 for GC–TOF data
When investigating specifically which metabolites occur only in the XA21 mediated response to PX099, i.e. signals that were specific to XA21/PXO99 but are not found in any of the other combinations, we found an unknown with the BinBase database identifier number 203611 (quantification m/z 128, FAME-based retention index 223571). The closest annotation was cyclohexylamine, but the identity was not further validated. In follow-up investigations, the hypothesis of an involvement of this tentatively annotated compound in the Xa21-mediated defense response needs to be examined.
3.6 Microarray analysis
Microarrays were used to profile gene expression in TP309 and Xa21 rice lines exposed to PXO99 or PXO99△raxST. Features showing less than 10% variation between replicate dye-swaps were used for analysis. Scanned electropherograms (Supplement S5) display the quality of total RNA in the pellet from 15 to 20 mg of rice leaf after metabolites had been solvent extracted. Discrete ribosomal RNA bands (represented as peaks in the electropherogram) were present in all samples, reflected by the presence of 18s and 28s RNA, with little indication of degradation. Figure 5 shows Volcano plots based on the differential abundances between pair-wise conditions, using fold-change and P-values for visualization. In an analogous way to LC–TOF data analysis, subsets of significantly differential genes were identified at P < 0.05 and fold-ratios >2.0. Clearly, the largest number of differentially expressed genes was between TP309_Xa21, mock-treated and PXO99-treated conditions (Fig. 5b). There were over 260 differentially expressed genes between TP309_Xa21_PXO99 and mock-treated; 39 between TP309_Xa21_PXO99△raxST and Mock-treated; and only 16 between TP309_PXO99 and mock-treated. Interestingly, this was over 15-fold higher number of differential genes between Xa21_PXO99 than the same comparison in TP309, and was corrected for the same amount of intact cRNA used in the hybridization. Similar differences in the number of responses were found for metabolite regulation (Table 3), indicating the importance of metabolism in pathogen responses. A summary table highlighting some of the significant differentially expressed genes is included in Supplement S6.
Gene expression differences in infected rice leaves. Volcano plot in Genespring-GX with P-values and intensity ratios (infected versus mock) as log-scaled axes. Significant differences at P < 0.05 with >2-fold intensity ratios are shown. Quality control for microarray data is given as Supplemental data S5 and Tables of differentially expressed genes are given as Supplemental information S6. a TP309_Mock versus TP309_PXO99. b TP309_Xa21-Mock versus TP309_Xa21-PXO99. c TP309_Xa21-Mock versus TP309_Xa21-PXO99△raxST
3.7 Correlating the results of transcriptomics and metabolomics provides several clues towards explaining which pathways are responsible for Xa21 resistance to PXO99
Next, we summarize the results of our profiling analyses by focusing on a few examples that highlight the challenges of interpreting two or more ‘omics’ profiling experiments (Table 4). Significantly differential genes were compared to the levels of metabolite products, associated with the corresponding enzymes, or to metabolite products along a particular pathway. For example, Isocitrate lyase (ICL1) was differential at the transcriptional level, but we were unable to correlate any significant change in levels of the associated substrate and product. Although the ICL1 transcript was induced in PXO99 compared to mock infected TP309, it was not differential in the Xa21 lines. LC–TOF analysis, however, revealed highly elevated levels of C6H8O7, likely to be citric acid or isocitrate, in both Xa21 lines but not in TP309. ICL1 catalyzes the formation of succinate and glyoxylate from isocitrate, a key reaction of the glyoxylate cycle most prominently known for germination, but also generally functioning at a branch point of carbon metabolism by diverting isocitrate through a carbon-conserving pathway, the glyoxylate cycle, thus preventing the oxidation steps in the TCA cycle. Expression of ICL1 is induced by growth on ethanol and repressed by growth on glucose (Fernandez et al. 1992; Luttik et al. 2000). In comparison to the upregulation of ICL1, glycolysis rates were found to be decreased in PXO99-challenged plants compared to mock-treated controls resulting in higher glucose and lower pyruvate levels (GC–TOF data Supplement S2). Consequently, pantothenic acid, an important co-factor for further oxidation of pyruvate to acetyl-CoA, was also found to be decreased in PXO99-challenged plants for both TP309 and Xa21 genotypes (Supplement S2).
Other over-expressed genes included β-1,3-glucanase and chitinase (Table 4) in PXO99 compared to mock infected Xa21. Conversely, these genes were suppressed in the PXO99 challenged susceptible strain versus TP309 mock. Both Chitinases and β-1,3-glucanases catalyze a broad series of substrates and products. Transgenic rice plants that contain one or more fungal disease-resistant gene(s) have previously been reported to control fungal diseases of rice (O’Toole 2001). Chitinases and Beta-1,3-glucanases are fungal cell wall degrading enzymes and can be found in plants and microbes. Genes encoding different chitinases and β-1,3-glucanases have previously been cloned and used to transform tobacco, resulting in increased survival relative to control plants in the presence of pathogenic fungi (Benhamou et al. 1993; Collinge et al. 1993). Indeed, β-1,3-Glucanases are commonly found throughout the plant kingdom and there is abundant evidence that they are involved in the defense mechanism of plants against pathogen infection, and have been proposed to be involved in physiological processes such as the stress response (Simmons 1994). Apparently these genes, in addition to being induced by fungi, are also induced in TP309_Xa21 in response to PXO99 but not in Xa21-PXO99△raxST.
Perhaps the most compelling observation was the comparison of transcriptomics data and metabolite data for the putative transcript for Glutamate decarboxylase (GAD), which codes for an enzyme that catalyzes the decarboxylation of glutamate to γ-aminobutyric acid (GABA) and CO2. The decrease of glutamate was confirmed by GC–TOF data, in accordance to the induction of GAD in both strains that were challenged with PXO99 and PXO99△raxST. GAD was particularly elevated in the Xa21 line (Table 4). LC–TOF analysis also revealed elevated levels for C4H9NO2. This empirical formula includes annotation as GABA in the METLIN database. An overlay of the EICs for the corresponding m/z revealed a clear increase in the abundance for Xa21_PXO99 samples relative to Xa21_mock (data not shown). Although the role of GABA in plants remains controversial (Bouche et al. 2004; Bouche and Fromm 2004; Bown et al. 2006), environmental perturbations of oxygen, water, and temperature, is reported to prompt the accumulation of GABA in plant cells (Shelp et al. 1999). GC–TOF data indicated that GABA might be furthered into cyclization to butyrolactam as outlined above, but the significance of this process for pathogen response is not established in literature.
Phenylalanine metabolism was significantly affected in PXO99 challenged XA21 rice variety, but not in the wild-type variety. Phenylalanine ammonia lyase (PAL), a key defense-related enzyme of the phenyl propanoid pathway, has recently been associated with disease resistance in rice plants via synthesis of isoflavones, phytoalexins, and lignin, compounds (Lin et al. 2008). Indeed rapid accumulation of PAL mRNA following inoculation with a pathogen has been described for maize (Cui et al. 2002). Moreover, rice leaves inoculated with Magnaporthe grisea significantly increased the enzyme activity of PAL (Cai et al. 2008). The PAL transcript in our study was significantly induced only in Xa21_PXO99 compared to Xa21_mock (Table 4; Supplement S5). Taken together, these results are consistent with our GC–TOF data for Phenylalanine where it was elevated.
A large number of glutathione and putative glutathione S-transferases (GST), as well as a glutathione transport protein were either detected or significantly induced in PX099 challenged relative to mock Xa21. In TP309, the induction/repression response to PXO99 was dependent on the GST––or putative GST––and on average, both in the magnitude and number of GSTs cancelled out any differences between PXO99 challenged and mock. The Xa21 line challenged with PXO99△raxST showed many GSTs to be significantly induced relative to mock. Interestingly, however, although several GSTs were detected, no significant differences were observed between Xa21_PXO99 compared to Xa21_Mock. GSTs detoxify endogenous compounds such as peroxidised lipids, they bind to toxins, or function as transport proteins, all of which might indicate a general response to stress. Oxidized glutathione levels were significantly different by LC–TOF analysis in the PXO99△raxST Xa21 line, detected but not altered in PXO99 challenged Xa21, and reduced in the wild-type TP309. This is an interesting result suggesting that PXO99 challenged Xa21 is not stressed and that the oxidized form of glutathione in response to PXO99 may be depleted in TP309 wild-type cells. Such hypotheses would need to be validated by target analyses of GSH/GSSG ratios.
Lipid Transfer Proteins (Ltp) reversibly bind and transport hydrophobic molecules in vitro and are likely to play a role in key processes of plant physiology. The induction of Ltp gene expression has been previously characterized in a fungus, and coincides with a transient increase in the expression levels of phenylalanine ammonia-lyase (PAL) gene, as well as salicylic acid (SA) gene in mycorrhizal roots (Blilou et al. 2000). Although we were able to detect induction of Ltp b1 transcripts in TP309 cells, we did not find it to correlate with co-expression or differential expression with either PAL or phenylalanine and salicylic acid levels in PXO99 challenged TP309 condition (Table 4). However, these metabolites were detected in PX099_Xa21 challenged plants instead and can be due to factors including that our system was non-fungal, limitations of assay sensitivity, as well as offsets between maximal expression of transcripts and metabolites.
4 Conclusions
An optimized sample processing protocol, starting from the same leaf sample of average weight of 15–20 mg FW, yielded sufficient material for parallel analysis by LC–TOF, GC–TOF and microarrays. This protocol is useful for investigation of complex processes such as plant infection and immunity, in leaf samples at different distances from the location of the infection. Unbiased, discovery-based metabolomics and transcriptomics analyses yielded novel insights into the rice response to Xoo. Our results reveal global metabolic and transcriptomic changes in leaf tissues of the XA21 transgenic variant challenged with PX099 but not to PXO99△raxST. While central carbon catabolism is reduced in correlation between gene and metabolite expression in both Xa21 and TP309 rice genotypes, genes and metabolic precursors for alkaloid biosynthesis are increased specifically in the XA21-mediated response to PX099 but not to PXO99△raxST. The outcome of ‘omics’ studies such as these will aid in a better understanding of complex response to pathogen infection (Tarpley et al. 2005). Although our analyses shows the robustness of our system, enabling differentiation of metabolite and transcript responses of two different rice genotypes and bacterial strains, coherent interpretation of transcriptomics and metabolomics results is very challenging, requiring carefully designed experiments, a large number of biological replicates and inclusion of time course studies to provide the appropriate context for further biological conclusions.
References
Benhamou, N., Broglie, K., et al. (1993). Antifungal effect of bean endochitinase on Rhizoctonia solani: Ultrastructural changes and cytochemical aspects of chitin breakdown. Canadian Journal of Microbiology, 39(3), 318–328.
Benjamini, Y., & Hochberg, Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. Journal of the Royal Statistical Society, Series B (Methodological), 57(1), 289–300.
Bera, S., & Purkayastha, R. P. (1999). Multicomponent coordinated defence response of rice to Rhizoctonia solani causing sheath blight. Current Science, 76(10), 1376–1384.
Blilou, I., Ocampo, J. A., et al. (2000). Induction of Ltp (lipid transfer protein) and Pal (phenylalanine ammonia-lyase) gene expression in rice roots colonized by the arbuscular mycorrhizal fungus Glomus mosseae. Journal of Experimental Botany, 51(353), 1969–1977.
Bouche, N., Fait, A., et al. (2004). The root-specific glutamate decarboxylase (GAD1) is essential for sustaining GABA levels in Arabidopsis. Plant Molecular Biology, 55(3), 315–325.
Bouche, N., & Fromm, H. (2004). GABA in plants: Just a metabolite? Trends in Plant Science, 9(3), 110–115.
Bown, A. W., Macgregor, K. B., et al. (2006). Gamma-aminobutyrate: Defense against invertebrate pests? Trends in Plant Science, 11(9), 424–427.
Brosche, M., Vinocur, B., et al. (2005). Gene expression and metabolite profiling of Populus euphratica growing in the Negev desert. Genome Biology, 6(12), R101.
Burdman, S., Shen, Y., et al. (2004). RaxH/RaxR: A two-component regulatory system in Xanthomonas oryzae pv. oryzae required for AvrXa21 activity. Molecular Plant-Microbe Interactions, 17(6), 602–612.
Cai, K., Gao, D., et al. (2008). Physiological and cytological mechanisms of silicon-induced resistance in rice against blast disease. Physiologia Plantarum, 134(2), 324–333.
Cho, K., Shibato, J., et al. (2008). Integrated transcriptomics, proteomics, and metabolomics analyses to survey ozone responses in the leaves of rice seedling. Journal of Proteome Research, 7(7), 2980–2998.
Collinge, D. B., Kragh, K. M., et al. (1993). Plant chitinases. Plant Journal, 3(1), 31–40.
Cui, Y., Magill, J., et al. (2002). Chalcone synthase and phenylalanine ammonia-lyase mRNA levels following exposure of sorghum seedlings to three fungal pathogens. Physiological and Molecular Plant Pathology, 49(3), 187–199.
da Silva, F. G., Shen, Y., et al. (2004). Bacterial genes involved in type I secretion and sulfation are required to elicit the rice Xa21-mediated innate immune response. Molecular Plant-Microbe Interactions, 17(6), 593–601.
Dharmadi, Y., & Gonzalez, R. (2004). DNA microarrays: Experimental issues, data analysis, and application to bacterial systems. Biotechnology Progress, 20(5), 1309–1324.
Eagle, H. (1959). Amino acid metabolism in mammalian cell cultures. Science, 130(3373), 432–437.
Eagle, H., Piez, K. A., et al. (1959). Protein turnover in mammaliar cell cultures. The Journal of Biological Chemistry, 234(3), 592–597.
Farrokhi, N., Burton, R. A., et al. (2006). Plant cell wall biosynthesis: Genetic, biochemical and functional genomics approaches to the identification of key genes. Plant Biotechnology Journal, 4(2), 145–167.
Fernandez, E., Moreno, F., et al. (1992). The ICL1 gene from Saccharomyces cerevisiae. European Journal of Biochemistry, 204(3), 983–990.
Fiehn, O., Kopka, J., et al. (2000). Metabolite profiling for plant functional genomics. Nature Biotechnology, 18(11), 1157–1161.
Fiehn, O., Wohlgemuth, G., et al. (2005). Setup and annotation of metabolomic experiments by integrating biological and mass spectrometric metadata. Proceedings of the Lecture Notes in Bioinformatics, 3615, 224–239.
Finkelstein, D., Ewing, R., et al. (2002). Microarray data quality analysis: Lessons from the AFGC project. Arabidopsis functional genomics consortium. Plant Molecular Biology, 48(1–2), 119–131.
Frey, I. M., Rubio-Aliaga, I., et al. (2007). Profiling at mRNA, protein, and metabolite levels reveals alterations in renal amino acid handling and glutathione metabolism in kidney tissue of Pept2−/− mice. Physiol Genomics, 28(3), 301–310.
Henikoff, S., & Comai, L. (2003). Single-nucleotide mutations for plant functional genomics. Annual Review of Plant Biology, 54, 375–401.
Hirai, M. Y., Yano, M., et al. (2004). Integration of transcriptomics and metabolomics for understanding of global responses to nutritional stresses in Arabidopsis thaliana. Proceedings of the National Academy of Sciences of the United States of America, 101(27), 10205–10210.
Hoefgen, R., & Nikiforova, V. J. (2008). Metabolomics integrated with transcriptomics: Assessing systems response to sulfur-deficiency stress. Physiologia Plantarum, 132(2), 190–198.
Jackson, M. J., Beaudet, A. L., et al. (1986). Mammalian urea cycle enzymes. Annual Review of Genetics, 20, 431–464.
Jayaraj, J., Yi, H., et al. (2004). Foliar application of Bacillus subtilis AUBS1 reduces sheath blight and triggers defense mechanisms in rice. Zeitschrift Fur Pflanzenkrankheiten Und Pflanzenschutz-Journal of Plant Diseases and Protection, 111(2), 115–125.
Jumtee, K., Bamba, T., et al. (2008). Integrated metabolite and gene expression profiling revealing phytochrome A regulation of polyamine biosynthesis of Arabidopsis thaliana. Journal of Experimental Botany, 59(6), 1187–1200.
Kaplan, F., Kopka, J., et al. (2007). Transcript and metabolite profiling during cold acclimation of Arabidopsis reveals an intricate relationship of cold-regulated gene expression with modifications in metabolite content. Plant Journal, 50(6), 967–981.
Kauffman, H., Reddy, A. P. K., Hsieh, S. P. V., & Marca, S. D. (1973). An improved technique for evaluation of resistance of rice varieties to Xanthomonas oryzae. Plant Disease Reporter, 57, 537–541.
Khan, W., Prithiviraj, B., et al. (2003). Chitosan and chitin oligomers increase phenylalanine ammonia-lyase and tyrosine ammonia-lyase activities in soybean leaves. Journal of Plant Physiology, 160(8), 859–863.
Kind, T., Wohlgemuth, G., et al. (2009). FiehnLib: Mass spectral and retention index libraries for metabolomics based on quadrupole and time-of-flight gas chromatography/mass spectrometry. Analytical Chemistry, 81(24), 10038–10048.
Lee, S. W., Han, S. W., et al. (2006). From the academy: Colloquium review. Unique characteristics of Xanthomonas oryzae pv. oryzae AvrXa21 and implications for plant innate immunity. Proceedings of the National Academy of Sciences of the United States of America, 103(49), 18395–18400.
Lee, S. W., Han, S. W., et al. (2009). A type I-secreted, sulfated peptide triggers XA21-mediated innate immunity. Science, 326(5954), 850–853.
Lin, Y. Z., Chen, H. Y., et al. (2008). Proteomic analysis of rice defense response induced by probenazole. Phytochemistry, 69(3), 715–728.
Luttik, M. A., Kotter, P., et al. (2000). The Saccharomyces cerevisiae ICL2 gene encodes a mitochondrial 2-methylisocitrate lyase involved in propionyl-coenzyme A metabolism. Journal of Bacteriology, 182(24), 7007–7013.
Matsuzaki, K., Kato, H., et al. (2005). Transcriptomics and metabolomics of dietary leucine excess. Journal of Nutrition, 135(6 Suppl), 1571S–1575S.
Mohanty, S. K., & Sridhar, R. (1982). Physiology of rice Tungro virus-disease––proline accumulation due to infection. Physiologia Plantarum, 56(1), 89–93.
Morris, S. M., Jr. (2007). Arginine metabolism: Boundaries of our knowledge. Journal of Nutrition, 137(6 Suppl 2), 1602S–1609S.
Nam, H., Chung, B. C., et al. (2009). Combining tissue transcriptomics and urine metabolomics for breast cancer biomarker identification. Bioinformatics, 25(23), 3151–3157.
O’Toole, J., Toenniessen, G., et al. (2001). The Rockefeller foundation’s international program on rice biotechnology.
Oliver, D. J., Nikolau, B., et al. (2002). Functional genomics: High-throughput mRNA, protein, and metabolite analyses. Metabolic Engineering, 4(1), 98–106.
Park, C. J., Peng, Y., et al. (2008). Rice XB15, a protein phosphatase 2C, negatively regulates cell death and XA21-mediated innate immunity. PLoS Biol, 6(9), e231.
Scholz, M., & Fiehn, O. (2007). SetupX––a public study design database for metabolomic projects. Pacific Symposium on Biocomputing, 12, 169–80.
Shelp, B. J., Bown, A. W., et al. (1999). Metabolism and functions of gamma-aminobutyric acid. Trends in Plant Science, 4(11), 446–452.
Shen, Y., Sharma, P., et al. (2002). The Xanthomonas oryzae pv. lozengeoryzae raxP and raxQ genes encode an ATP sulphurylase and adenosine-5′-phosphosulphate kinase that are required for AvrXa21 avirulence activity. Molecular Microbiology, 44(1), 37–48.
Simmons, C. R. (1994). The physiology and molecular biology of plant 1,3-ß-glucanases and 1,3;1,4-ß-glucanases. Critical Reviews in Plant Sciences, 13, 325–387.
Song, W. Y., Wang, G. L., et al. (1995). A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science, 270(5243), 1804–1806.
Spencer, P. A., & Towers, G. H. N. (1991). Restricted occurrence of acetophenone signal compounds. Phytochemistry, 30, 2933–2937.
Steinberg, C. E., Sturzenbaum, S. R., et al. (2008). Genes and environment––striking the fine balance between sophisticated biomonitoring and true functional environmental genomics. Science of the Total Environment, 400(1–3), 142–161.
Tan, K. C., Ipcho, S. V., et al. (2009). Assessing the impact of transcriptomics, proteomics and metabolomics on fungal phytopathology. Mol Plant Pathol, 10(5), 703–715.
Tarpley, L., Duran, A. L., et al. (2005). Biomarker metabolites capturing the metabolite variance present in a rice plant developmental period. BMC Plant Biol, 5, 8.
The International Rice Genome Sequencing Project. (2005). The map-based sequence of the rice genome. Nature, 436(7052), 793–800.
Trung-Nghia, P., Bassie, L., et al. (2003). Reduction in the endogenous arginine decarboxylase transcript levels in rice leads to depletion of the putrescine and spermidine pools with no concomitant changes in the expression of downstream genes in the polyamine biosynthetic pathway. Planta, 218(1), 125–134.
Tukey, J. W. (1949). Comparing individual means in the analysis of variance. Biometrics, 5, 99.
Urbanczyk-Wochniak, E., Luedemann, A., et al. (2003). Parallel analysis of transcript and metabolic profiles: A new approach in systems biology. EMBO Reports, 4(10), 989–993.
Verhoeckx, K. C., Bijlsma, S., et al. (2004). Characterization of anti-inflammatory compounds using transcriptomics, proteomics, and metabolomics in combination with multivariate data analysis. International Immunopharmacology, 4(12), 1499–1514.
Vidhyasekaran, P., Kamala, N., et al. (2001). Induction of systemic resistance by Pseudomonas fluorescens Pf1 against Xanthomonas oryzae pv. oryzae in rice leaves. Phytoparasitica, 29(2), 155–166.
Vijayendran, C., Barsch, A., et al. (2008). Perceiving molecular evolution processes in Escherichia coli by comprehensive metabolite and gene expression profiling. Genome Biology, 9(4), R72.
Weckwerth, W., Wenzel, K., et al. (2004). Process for the integrated extraction, identification and quantification of metabolites, proteins and RNA to reveal their co-regulation in biochemical networks. Proteomics, 4(1), 78–83.
Yang, S., Tschaplinski, T. J., et al. (2009). Transcriptomic and metabolomic profiling of Zymomonas mobilis during aerobic and anaerobic fermentations. BMC Genomics, 10, 34.
Acknowledgments
This work was financially supported by NSF grant MCB-0820823 to OF and NIH grant GM55962 to PCR and DOE grant # DE-AC02-05CH11231 to PCR. Technical assistance by Yun Lu for GC–TOF data acquisition is appreciated. The assistance of Sean Coughlan in supplying the Agilent rice microarrays and Kshama Mehta who helped perform some of the microarray hybridizations is appreciated.
Authors’ contributions
TRS carried out microarray and LC–TOF analyses, data interpretation and writing the manuscript. SF performed LC–TOF and data analysis. AK assisted with sample extraction and preparation. KHJ performed the bacterial challenge tests on the various rice genotypes. SC provided the rice arrays, KM assisted with microarray hybridization. GW altered BinBase algorithms to retain only high quality GC–TOF spectra for rice metabolites. OF performed GC–TOF data analysis, data interpretation and writing the manuscript. PR helped plan the experiments and assisted with interpretation of results and writing of the MS.
Competing interests
The author(s) declare that they have no competing interests.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Sana, T.R., Fischer, S., Wohlgemuth, G. et al. Metabolomic and transcriptomic analysis of the rice response to the bacterial blight pathogen Xanthomonas oryzae pv. oryzae . Metabolomics 6, 451–465 (2010). https://doi.org/10.1007/s11306-010-0218-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11306-010-0218-7