Abstract
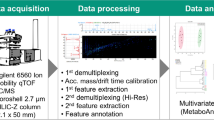
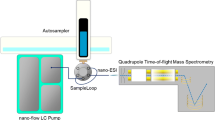
With unmatched mass resolution, mass accuracy, and exceptional detection sensitivity, Fourier Transform Ion Cyclotron Resonance Mass Spectrometry (FTICR-MS) has the potential to be a powerful new technique for high-throughput metabolomic analysis. In this study, we examine the properties of an ultrahigh-field 12-Tesla (12T) FTICR-MS for the identification and absolute quantitation of human plasma metabolites, and for the untargeted metabolic fingerprinting of inbred-strain mouse serum by direct infusion (DI). Using internal mass calibration (mass error ≤1 ppm), we determined the rational elemental compositions (incorporating unlimited C, H, N and O, and a maximum of two S, three P, two Na, and one K per formula) of approximately 250 out of 570 metabolite features detected in a 3-min infusion analysis of aqueous extract of human plasma, and were able to identify more than 100 metabolites. Using isotopically-labeled internal standards, we were able to obtain excellent calibration curves for the absolute quantitation of choline with sub-pmol sensitivity, using 500 times less sample than previous LC/MS analyses. Under optimized serum dilution conditions, chemical compounds spiked into mouse serum as metabolite mimics showed a linear response over a 600-fold concentration range. DI/FTICR-MS analysis of serum from 26 mice from 2 inbred strains, with and without acute trichloroethylene (TCE) treatment, gave a relative standard deviation (RSD) of 4.5%. Finally, we extended this method to the metabolomic fingerprinting of serum samples from 49 mice from 5 inbred strains involved in an acute alcohol toxicity study, using both positive and negative electrospray ionization (ESI). Using these samples, we demonstrated the utility of this method for high-throughput metabolomics, with more than 400 metabolites profiled in only 24 h. Our experiments demonstrate that DI/FTICR-MS is well-suited for high-throughput metabolomic analysis.






Similar content being viewed by others
References
Aharoni, A., Ric de Vos, C. H., Verhoeven, H. A., Maliepaard, C. A., Kruppa, G., Bino, R., Goodenowe, D. B. (2002). Nontargeted metabolome analysis by use of Fourier transform ion cyclotron mass spectrometry, Omics, 6(3), 217–234.
Allen, J., Davey, H. M., Broadhurst, D., Heald, J. K., Rowland, J. J., Oliver, S. G., Kell, D. B. (2003). High-throughput classification of yeast mutants for functional genomics using metabolic footprinting, Nature Biotechnology, 21(6), 692–696.
Belov, M. E., Anderson, G. A., Angell, N. H., Shen, Y., Tolic, N., Udseth, H. R., Smith, R. D. (2001). Dynamic range expansion applied to mass spectrometry based on data-dependent selective ion ejection in capillary liquid chromatography Fourier transform ion cyclotron resonance for enhanced proteome characterization, Analytical Chemistry, 73(21), 5052–5060.
Brown, S. C., Kruppa, G., Dasseux, J. L. (2005). Metabolomics applications of FT-ICR mass spectrometry, Mass Spectrometry Reviews, 24(2), 223–231.
Buchholz, A., Takors, R., Wandrey, C., (2001). Quantification of intracellular metabolites in Escherichia coli K12 using liquid chromatographic-electrospray ionization tandem mass spectrometric techniques, Analytical Biochemistry, 295(2), 129–137.
De Vos, R. C., Moco, S., Lommen, A., Keurentjes, J. J., Bino, R. J., Hall, R. D. (2007). Untargeted large-scale plant metabolomics using liquid chromatography coupled to mass spectrometry, Nature Protocols, 2(4), 778–791.
Denkert, C., Budczies, J., Kind, T., Weichert, W., Tablack, P., Sehouli, J., Niesporek, S., Konsgen, D., Dietel, M., Fiehn, O., (2006). Mass spectrometry-based metabolic profiling reveals different metabolite patterns in invasive ovarian carcinomas and ovarian borderline tumors, Cancer Research, 66(22), 10795–10804.
Dunn, W. B., Bailey, N. J., Johnson, H. E. (2005). Measuring the metabolome: Current analytical technologies, Analyst, 130(5), 606–625.
Eisen, M. B., Spellman, P. T., Brown, P. O., Botstein, D., (1998). Cluster analysis and display of genome-wide expression patterns, Proceedings of the National Academy of Sciences, 95(25), 14863–14868.
Fahy, E., Sud, M., Cotter, D., Subramaniam, S., (2007). LIPID MAPS online tools for lipid research, Nucleic Acids Research, 35(Web Server issue), W606–W612.
Fan, T. W., Lane, A. N., Higashi, R. M. (2004). The promise of metabolomics in cancer molecular therapeutics, Current Opinion in Molecular Therapeutics, 6(6), 584–592.
Fiehn, O., Kloska, S., Altmann, T., (2001). Integrated studies on plant biology using multiparallel techniques, Current Opinion in Biotechnology, 12(1), 82–86.
Galinski, E. A. (1995). Osmoadaptation in bacteria, Advances in Microbial Physiology, 37, 272–328.
Gamache, P. H., Meyer, D. F., Granger, M. C., Acworth, I. N. (2004). Metabolomic applications of electrochemistry/mass spectrometry, American Society for Mass Spectrometry, 15(12), 1717–1726.
Goodacre, R., Vaidyanathan, S., Dunn, W. B., Harrigan, G. G., Kell, D. B. (2004). Metabolomics by numbers: Acquiring and understanding global metabolite data, Trends in Biotechnology, 22(5), 245–252.
Halket, J. M., Przyborowska, A., Stein, S. E., Mallard, W. G., Down, S., Chalmers, R. A. (1999). Deconvolution gas chromatography/mass spectrometry of urinary organic acids–potential for pattern recognition and automated identification of metabolic disorders, Rapid Communications in Mass Spectrometry, 13(4), 279–284.
Hirai, M. Y., Yano, M., Goodenowe, D. B., Kanaya, S., Kimura, T., Awazuhara, M., Arita, M., Fujiwara, T., Saito, K., (2004). Integration of transcriptomics and metabolomics for understanding of global responses to nutritional stresses in Arabidopsis thaliana, Proceedings of the National Academy of Sciences, 101(27), 10205–10210.
Hughey, C. A., Rodgers, R. P., Marshall, A. G. (2002). Resolution of 11,000 compositionally distinct components in a single electrospray ionization Fourier transform ion cyclotron resonance mass spectrum of crude oil, Analytical Chemistry, 74(16), 4145–4149.
Jones, O. A., Spurgeon, D. J., Svendsen, C., Griffin, J. L. (2007). A metabolomics based approach to assessing the toxicity of the polyaromatic hydrocarbon pyrene to the earthworm Lumbricus rubellus. Chemosphere [Epub ahead of print].
Kind, T., Fiehn, O., (2006). Metabolomic database annotations via query of elemental compositions: Mass accuracy is insufficient even at less than 1 ppm, BMC Bioinformatics, 7, 234.
Kind, T., Fiehn, O., (2007). Seven Golden Rules for heuristic filtering of molecular formulas obtained by accurate mass spectrometry, BMC Bioinformatics, 8, 105.
Koc, H., Mar, M. H., Ranasinghe, A., Swenberg, J. A., Zeisel, S. H. (2002). Quantitation of choline and its metabolites in tissues and foods by liquid chromatography/electrospray ionization-isotope dilution mass spectrometry, Analytical Chemistry, 74(18), 4734–4740.
Kolch, W., Mischak, H., Pitt, A. R. (2005). The molecular make-up of a tumour: Proteomics in cancer research, Clinical Science (London), 108(5), 369–383.
Lee, S. H., Woo, H. M., Jung, B. H., Lee, J., Kwon, O. S., Pyo, H. S., Choi, M. H., Chung, B. C. (2007). Metabolomic approach to evaluate the toxicological effects of nonylphenol with rat urine, Analytical Chemistry, 79(16), 6102–6110.
Marshall, A. G., Hendrickson, C. L., Shi, S. D. (2002). Scaling MS plateaus with high-resolution FT-ICRMS, Analytical Chemistry, 74(9), 252A–259A.
Nakamura, Y., Kimura, A., Saga, H., Oikawa, A., Shinbo, Y., Kai, K., Sakurai, N., Suzuki, H., Kitayama, M., Shibata, D., Kanaya, S., & Ohta, D., (2007). Differential metabolomics unraveling light/dark regulation of metabolic activities in Arabidopsis cell culture. Planta, 227(1), 57–66.
Neubauer, O., (1901). Archiv, Fur Experimentelle Pathologie und Pharmakologie, 46, 133–154.
Nicholson, J. K., Lindon, J. C., Holmes, E., (1999). ‘Metabonomics’: Understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data, Xenobiotica, 29(11), 1181–1189.
Ogata, H., Goto, S., Sato, K., Fujibuchi, W., Bono, H., Kanehisa, M., (1999). KEGG: Kyoto Encyclopedia of Genes and Genomes, Nucleic Acids Research, 27(1), 29–34.
Ohta, D., Shibata, D., Kanaya, S., (2007). Metabolic profiling using Fourier-transform ion-cyclotron-resonance mass spectrometry, Analytical and Bioanalytical Chemistry, 389(5), 1469–1475.
Plumb, R., Castro-Perez, J., Granger, J., Beattie, I., Joncour, K., Wright, A., (2004). Ultra-performance liquid chromatography coupled to quadrupole-orthogonal time-of-flight mass spectrometry. Rapid Communications in Mass Spectrometry, 18(19), 2331–2337.
Pomfret, E. A., daCosta, K. A., Schurman, L. L., Zeisel, S. H. (1989). Measurement of choline and choline metabolite concentrations using high-pressure liquid chromatography and gas chromatography-mass spectrometry. Analytical Biochemistry, 180(1), 85–90.
Saldanha, A. J. (2004). Java Treeview–extensible visualization of microarray data, Bioinformatics, 20(17), 3246–3248.
Sato, S., Soga, T., Nishioka, T., Tomita, M., (2004). Simultaneous determination of the main metabolites in rice leaves using capillary electrophoresis mass spectrometry and capillary electrophoresis diode array detection, Plant Journal, 40(1), 151–163.
Soga, T., Baran, R., Suematsu, M., Ueno, Y., Ikeda, S., Sakurakawa, T., Kakazu, Y., Ishikawa, T., Robert, M., Nishioka, T., Tomita, M., (2006). Differential metabolomics reveals ophthalmic acid as an oxidative stress biomarker indicating hepatic glutathione consumption, Journal of Biological Chemistry, 281(24), 16768–16776.
Southam, A. D., Payne, T. G., Cooper, H. J., Arvanitis, T. N., Viant, M. R. (2007). Dynamic range and mass accuracy of wide-scan direct infusion nanoelectrospray Fourier transform ion cyclotron resonance mass spectrometry-based metabolomics increased by the spectral stitching method, Analytical Chemistry, 79(12), 4595–4602.
Staack, R. F., Varesio, E., Hopfgartner, G., (2005). The combination of liquid chromatography/tandem mass spectrometry and chip-based infusion for improved screening and characterization of drug metabolites, Rapid Communications in Mass Spectrometry, 19(5), 618–626.
Sud, M., Fahy, E., Cotter, D., Brown, A., Dennis, E. A., Glass, C. K., Merrill, A. H., Jr., Murphy, R. C., Raetz, C. R., Russell, D. W., Subramaniam, S., (2007). LMSD: LIPID MAPS structure database. Nucleic Acids Research, 35(Database issue), D527–D532.
Tohge, T., Nishiyama, Y., Hirai, M. Y., Yano, M., Nakajima, J., Awazuhara, M., Inoue, E., Takahashi, H., Goodenowe, D. B., Kitayama, M., Noji, M., Yamazaki, M., Saito, K., (2005). Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor, Plant Journal, 42(2), 218–235.
Want, E. J., O’Maille, G., Smith, C. A., Brandon, T. R., Uritboonthai, W., Qin, C., Trauger, S. A., Siuzdak, G., (2006). Solvent-dependent metabolite distribution, clustering, and protein extraction for serum profiling with mass spectrometry, Analytical Chemistry, 78(3), 743–752.
Wikoff, W. R., Gangoiti, J. A., Barshop, B. A., Siuzdak, G., (2007). Metabolomics identifies perturbations in human disorders of propionate metabolism. Clinical Chemistry. [Epub ahead of print].
Wishart, D. S., Tzur, D., Knox, C., Eisner, R., Guo, A. C., Young, N., Cheng, D., Jewell, K., Arndt, D., Sawhney, S., Fung, C., Nikolai, L., Lewis, M., Coutouly, M. A., Forsythe, I., Tang, P., Shrivastava, S., Jeroncic, K., Stothard, P., Amegbey, G., Block, D., Hau, D. D., Wagner, J., Miniaci, J., Clements, M., Gebremedhin, M., Guo, N., Zhang, Y., Duggan, G. E., Macinnis, G. D., Weljie, A. M., Dowlatabadi, R., Bamforth, F., Clive, D., Greiner, R., Li, L., Marrie, T., Sykes, B. D., Vogel, H. J., Querengesser, L., (2007). HMDB: The human metabolome database. Nucleic Acids Research, 35 (Database issue), D521–D526.
Acknowledgements
The authors would like to thank Dr. David Robinette for his assistance with the preparation of the manuscript. This study was partially funded by a gift from an anonymous donor to support research in proteomics at UNC, by NIH grants P30 CA0-16086, 1S10RR16776, caBIG-ICR-07-20-01, R01 AA016258, and P30 Es010126, by collaboration with Bruker Daltonics, and by start-up funds from the University of Victoria. We also acknowledge the use of the 12-Tesla FTICR mass spectrometer at the UNC-Duke Michael Hooker Proteomics Center, NC (NIH 1-S10-RR019889-01 and NCBC 2005-IDG-1015) for some of these experiments.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Han, J., Danell, R.M., Patel, J.R. et al. Towards high-throughput metabolomics using ultrahigh-field Fourier transform ion cyclotron resonance mass spectrometry. Metabolomics 4, 128–140 (2008). https://doi.org/10.1007/s11306-008-0104-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11306-008-0104-8