Abstract
With the increasing use of metabolomics as a means to study a large number of different biological research questions, there is a need for a minimal set of reporting standards that allow the scientific community to evaluate, understand, repeat, compare and re-investigate metabolomics studies. Here we propose, a first draft of minimal requirements to effectively describe the biological context of metabolomics studies that involve microbial or in vitro biological subjects. This recommendation has been produced by the microbiology and in vitro biology working subgroup of the Metabolomics Standards Initiative in collaboration with the yeast systems biology network as part of a wider standardization initiative led by the Metabolomics Society. Microbial and in vitro biology metabolomics is defined by this sub-working group as studies with any cell or organism that require a defined external medium to facilitate growth and propagation. Both a minimal set and a best practice set of reporting standards for metabolomics experiments have been defined. The minimal set of reporting standards for microbial or in vitro biology metabolomics experiments includes those factors that are specific for metabolomics experiments and that critically determine the outcome of the experiments. The best practice set of reporting standards contains both the factors that are specific for metabolomics experiments and general aspects that critically determine the outcome of any microbial or in vitro biological experiment.
Similar content being viewed by others
1 Introduction
Microbiology is arguably one of the oldest sciences going back thousands of years. In brewing, people had added relatively well-defined mixtures of components to microorganisms in batch culture in order to produce characteristic flavors of beverages or to manipulate alcohol content. The science of microbiology/in vitro biology has subsequently evolved to the study of organism/cell responses in precise treatments to minimize complications arising from variable environmental influences. As with any biological system, metabolomics strives to unravel in a non-targeted and comprehensive manner the metabolic behavior of a system to an environmental condition or perturbation. Current applications of metabolomics to microbiology focuses on the area of microbial production, where the goal is to identify bottlenecks in biosynthesis routes in order to increase the flux from substrate to product (van der Werf 2005). In the field of in vitro biology, metabolomics is used as a means to obtain a comprehensive view of the reaction of a cell to specific toxic or pharmaceutical compounds. Further applications of metabolomics in both microbiology and in vitro biology include the identification of novel (bioactive) compounds, identification of critical medium compounds, the characterization of particular cellular metabolic phenotypes, understanding of cell physiology and its response to for instance stress or loss of homeostasis, dissecting the biochemical interaction among microbial communities, and others. In combination with other ‘omics’ technologies such as transcriptomics, proteomics or fluxomics, metabolomics is receiving increasing interest in systems oriented approaches aiming at a comprehensive understanding of the biological system as a whole (Bruggeman and Westerhoff 2006).
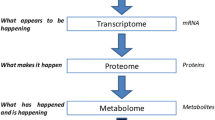
Metabolomics, like the other functional genomics technologies, is a complex technology which comprises many different steps (Fig. 1) ultimately resulting in large data sets. However, metabolomics is not about data generation, but about translating metabolome data into biologically relevant information. To this end, it is essential that representative ‘snapshot’ metabolomes are generated of which the metabolite composition is identical to the metabolome composition of the cells at the time of harvesting (Villas Boas et al. 2005). In this respect, the analysis of metabolomes is much more challenging than that of transcriptomes and proteomes as the turnover of metabolites is much higher: in the order of milliseconds to seconds compared to minutes and hours for mRNAs and proteins, respectively. Moreover, a metabolome data set and associated metadata should contain sufficient information in order to be able to answer the biological question under study and therefore experimental design (Trygg et al. 2006) is another key aspect of metabolomics studies.
In view of this, it is essential for the scientific community to be able to judge the quality of the results described in metabolomics papers that are starting to appear. To this end, a set of minimal reporting standards for scientist to be able to evaluate, understand, repeat, compare and re-investigate metabolomics data are of key importance.
2 The standards generation process
2.1 Organization
Under auspices of an oversight committee of the Metabolomics Society (http://www.metabolomicssociety.org/mstandards.html), different workgroups were formulated that together form the Metabolomics Standards Initiative (MSI) (http://www.msi-workgroups.sourceforge.net/). These working groups include biological sample context, chemical analysis, data analysis, ontology and data exchange (see also reports of these working groups in this same issue of Metabolomics).
2.2 Approach; the standards generation process
Standards reported by other functional genomics communities such as MIAMI (Minimum Information About a Microarray Experiment—Brazma et al. 2001), RSBI (Reporting Structure for Biological Investigations—http://www.mged.org/workgroups/rsbi/rsbi.html), FuGO (Functional Genomics Ontology—http://www.obi.sourceforge.net/), MIGS (Minimum information about a genome sequence—Field et al. 2006), CEBS-DD (Chemical effects in biological systems—data dictionary—Fostel et al. 2005), and PSI (The HUPO Proteomics Standards Initiative—Taylor 2006), and initial attempts to come to reporting standards for metabolomics experiments SMRS (Standard metabolic reporting structures—Lindon et al. 2005) and ARMET (Architecture for metabolomics—Jenkins et al. 2004 and 2005) were evaluated. These were used as the input for coming to the metabolomics reporting standards reported here.
The microbiology and in vitro biology standards were generated in a sub-working group of the biological sample context workgroup of the metabolomics standards initiative under auspices of the metabolomics society (http://www.metabolomicssociety.org) in collaboration with the yeast systems biology network (http://www.ysbn.org). The biological sample context working group deals with aspects starting from defining the exact biological question to experimental design to sample generation and sample work-up (see also Fig. 1). Sample work-up was in part also covered by the chemical analysis working group as both biological and analytical aspects are important in working up samples that are representative for the biological situation that they were derived from. The draft version of these for microbiology and in vitro biology metabolomics reporting standards as proposed by these authors were presented at the 2nd scientific meeting of the Metabolomics Society (Boston, 24–29 June 2006), the Metabomeeting3 (London, 18–19 December 2006) and two Yeast Systems Biology Network Meetings (Helsinki, 22 June 2006 and Gossau, 9 June 2007), and the input received at these meetings has resulted in the reporting standards proposed in this paper.
3 Standards
3.1 Ontology
In this manuscript no specific attention was paid to ontology. The terminology used is that general to the fields of microbiology and in vitro biology. Our terminology requirements and recommendations will be collected by the MSI Ontology working group (Sansone et al. this issue), operating under the Open Biomedical Ontology (OBO, http://www.obo.sourceforge.net) umbrella.
3.2 Microbial and in vitro biology metabolomics
Microbial and in vitro biology metabolomics is defined by this sub-working group as studies with any cell or organism that require a defined external medium to facilitate growth and propagation.
3.3 Scope of this recommendation
The scope the sub-working group on microbial and in vitro biology sample context was to identify, develop and disseminate best reporting practices in all aspects of microbial and in vitro biology metabolomics that are related to describing the samples generated in microbial or in vitro biological studies to be analyzed by metabolomics. The focus of the efforts is on metabolomics-specific methodologies and/or technical information that are critical to the outcome of metabolomics experiments.
3.4 Aim
The aim was not be to prescribe how to perform a microbial or in vitro biology metabolomics experiment, but to formulate a minimum set of reporting standards that describe the methods (what are the methods and how they were actually executed). Consequently, there is no attempt to restrict or dictate specific practices, but to develop consistent and appropriate descriptors to allow the evaluation of the experiments performed and to support the dissemination and re-use of metabolomics data.
3.5 Proposed reporting standards on microbial and in vitro biology sample context: starting point
The metabolomics standards for reporting on samples in in vitro biology or microbial metabolomics experiments builds upon the general biological practice of reporting biological experiments in a way that the materials and methods section should include ‘sufficient, but brief, technical information to allow the experiments to be repeated’ as described in the instructions to authors, of journals like the Journal of Biological Chemistry (http://www.jbc.org), Microbiology-UK (http://www.mic.sgmjournals.org) or the journals of the American Society of Microbiology (http://www.journals.asm.org/ ASM). This includes aspects like:
-
Species/strains/bioresource
-
Source of the strains and substrates
-
Experimental design
-
Inoculation procedure
-
Growth and/or treatment conditions
-
Time of sampling and of other events relevant to the samples
These aspects were considered to be general aspects that are reported in every biological scientific paper/experiment, and are not a part of the minimal set of reporting standards (Sect. 3.6) as identified by this sub-working group. However, this does not mean that these factors are not critical to the outcome of a metabolomics experiment, and therefore also a best practice set of reporting standards (Sect. 3.7) for reporting microbial or in vitro biology metabolomics experiments was defined.
3.6 Proposed minimal set of reporting standards for microbial and in vitro biology metabolomics experiments
The minimal set of reporting standards for in vitro biology or microbial metabolomics experiments includes those factors that are specific for metabolomics experiments and that critically determine the outcome of the experiments. These metabolomics-specific factors should be described in larger detail. As metabolite data are extremely sensitive to environmental conditions, also the cultivation conditions and harvesting time points, aspects general to microbial and in vitro biology studies, should be reported as accurately as possible.
Metabolomics-specific factors that together form the minimal reporting standards for microbial and in vitro biology metabolomics experiments are:
-
Sampling
-
What is the time between sample removal from its environment until metabolic activity is truly stopped?
-
What was the temperature during this process?
-
How were the samples harvested?
-
-
Quenching
-
How was the metabolism of the samples shut down?
-
How is the cell integrity under the quenching conditions?
-
-
Extracellular metabolites
-
How were intracellular metabolites discriminated from extracellular metabolites?
-
-
Extraction of metabolites from the cells
-
How were the (intracellular) metabolites extracted from the cells?
-
What is the estimated recovery at this step?
-
-
Normalization of the metabolome data
-
How were the metabolome data normalized?
-
Specifically (in case of normalizing the data with respect to the amount of cells (no., mg) that they were obtained from): How and at what step was the amount of biomass determined?
-
-
-
Sample clean-up/work-up
-
How were the samples cleaned-up with respect to compounds that interfere with analysis?
-
-
Sample storage
-
How and how long were the samples stored after collection, during work-up and prior to analysis?
-
-
Quality control steps
-
How was verified that no biotic or a-biotic changes occurred, or were at least minimized, during the complete sample collection and work-up phase?
-
-
Detection limit
-
What is the detection limit of the metabolites for the samples analyzed in the study?
-
-
Stability
-
What is known about the stability of (specific) metabolites during quenching, extraction and sample preprocessing?
-
In view of the many steps in a metabolomics experiment, we also advise to include a full schedule of the sample processing and analysis protocol.
3.7 Best-practice set of reporting standards for microbial and in vitro biology metabolomics experiments
The best practice set of reporting standards for microbial and in vitro biology metabolomics experiments contains both the factors that are specific for metabolomics experiments (see minimal set of reporting standards—Sect. 3.6) and general aspects that critically determine the outcome of any in vitro biological or microbiological experiments.
General factors that critically determine the outcome of any microbial and in vitro biology experiments are:
-
Experimental design
-
Biological question/the goal of the experiment
-
Experimental design (preferably in the form of a diagram or table)—relationships between:
-
Samples
-
Treatments/growth conditions
-
Time/timing of sample collection
-
Extracts
-
Repeats
-
Metabolome samples
-
Phenotypic characteristics of the samples
-
Other ‘omics’ data sets generated from these same samples
-
-
-
Biosource
-
Source/supplier of the cell line/strain (e.g. ATCC,...)
-
In case of natural isolates:
-
If at all possible, cultures should be deposited in an international culture collection with an accession number and relevant details
-
As micro-organisms are often identified incorrectly: What taxonomic system was used to identify the (micro-)organism?
-
-
In case of mutant strains:
-
From which wild-type were they obtained and how?
-
-
In case of (higher) eukaryotes
-
Cell type, organ derivation, grade of differentiation, subcellular location
-
Immortalized or transformed (if applicable)
-
-
Cell storage
-
-
Growth environment
-
Growth container: Type, supplier, geometry of the fermenter/bioreactor, (shake) flask or microtiter plates
-
Growth supports (type and supplier) in case of cells cultured in adherence
-
-
Growth configuration (suspension or attached culture, monolayer, double layer, sandwich, spheroids, batch, fed-batch, perfusion, continuous fermentation,...)
-
Inoculation procedure
-
Subculturing and splitting protocols
-
Inoculation size, seeding density (volume% [v/v], n° of cells/ml for suspension cultures; n° of cells/cm2 for cells in adherence, subconfluence or confluence, ...)
-
-
Medium/substrates (type and supplier, concentration/percentage)—including additions and supplementations (antibiotics, growth factors, serum type, and batches, concentration/percentage, ...)
-
Environmental conditions: Temperature, pH, gas composition, humidity, % CO2, stirrer speed, evaporation, pO2
-
Which of the environmental conditions were controlled and which could alter (freely) during growth?
-
-
Growth rate
-
If the cells were grown at a set fixed growth rate: which was the growth rate?
-
-
-
Treatment/incubation conditions
-
Treatment factors
-
Biotic (e.g. competition with or infection by other organisms)
-
Abiotic (e.g. physical stresses, chemical substances, ...)
-
-
Treatment dose, vehicle
-
Pre-treatment and/or treatment time and intervals
-
In case of use/incubation with labeled substrates: 13C compounds used (% enrichment, purity), labeling protocols
-
Pretreatment if any
-
-
Harvesting
-
Biotic characteristics of moment of harvesting: growth phase/stage (logarithmic, stationary, steady state, cytostatic phase, cell cycle phase, ...), number of generations in case of continuous cultures, time of sampling, stabilization time/phase before experiment, number of culture passages, independent indicators of differentiated state (immunological or molecular markers)
-
A-biotic characteristics at time of harvesting (cell density [OD, DWT, counts], depletion of nutrients, treatment time)
-
Phenotypic characteristics especially relating to the question under study (i.e. yield, productivity, color, form, ...)
-
Aspects mentioned under minimal reporting standards (Sect. 3.6)
-
-
Biotic factors related to sample work up
-
Aspects mentioned under minimal reporting standards (Sect. 3.6)
-
4 Request for feedback
These reporting standards are a first attempt and feedback is encouraged. A checklist of the standards reported in this document has been registered at the MSI Portal (http://www.msi-workgroups.sourceforge.net/), a ‘one-stop shop’ of extant projects with the goal of fostering collaborative development and ultimately promoting gradual integration of functional genomics standards. For your comments, suggestions, additions, etc. on these standards please contact the chair of the MSI Working Subgroup on microbiology and in vitro biology sample context, Mariët J. van der Werf (mariet.vanderwerf@tno.nl or Msi-workgroups-feedback@lists.sourceforge.net).
5 Discussion
With the above described minimal set and best practice set of reporting standards we think that it should be possible to be able for the scientific community to judge the merits of the biological sample context part of a microbial and in vitro biology metabolomics study. A major issue in the discussions of the sub-working group on microbial and in vitro biology sample context was the amount of detail that should be included in the reporting standards. Some of the experts felt that every biological parameter that is important for generating a representative metabolomics sample should be covered in these reporting standards, while others were of the opinion that only those aspects that are specific to metabolomics experiments should be covered by the metabolomics reporting standards. Therefore, both a minimal set of reporting standards and a best practice set of reporting standards were defined.
We believe that with the division of the labor of biological sample workgroup of the metabolomics standards initiative into four sub-groups (in vivo/mammalian biology; plant biology; in vitro/microbiology; environmental—see also the contributions of these sub-working groups in this issue of Metabolomics) has ensured that each of the domains has received appropriate representation from their respective communities. Although several aspects are common to the four different biological sample context sub-working groups, a key difference between microbial and in vitro biology metabolomics experiments and metabolomics experiments from the other biological domains is that in microbial and in vitro biology metabolomics care should be taken to distinguish between cellular and added compounds. Ultimately, it is hoped that the efforts of the groups will come together to form a unified set of reporting requirements to represent the ‘biological context of metabolomics experiments’. Furthermore, given the growing number of minimal reporting standard initiatives coming from the other functional genomics domains, we wish that ultimately minimal reporting standards can be formulated that allow the description of the biological sample context in an ‘omic’-independent manner.
References
Brazma, A., Hingamp, P., Quackenbush, J., Sherlock, G., Spellman, P., Stoeckert, C., Aach, J., Ansorge, W., Ball, C. A., Causton, H. C., Gaasterland, T., Glenisson, P., Holstege, F. C. P., Kim, I. F., Markowitz, V., Matese, J. C., Parkinson, H., Robinson, A., Sarans, U., Schylze-Kremer, S., Stewart, J., Taylor, R., Violo, J., & Vingron, M. (2001). Minimum information about a microarray experiment (MIAME)—towards standards for microarray data. Nature Genetics, 29, 365–371.
Bruggeman, F. J., & Westerhoff, H.,V. (2006). The nature of systems biology. Trends in Microbiology, 15, 45–50.
Field, D., Morrison, N., Selengut, J., & Sterk, P. (2006). eGenomics: cataloguing our complete genome collection. Omics: A Journal of Integrative Biology, 10, 100–104.
Fostel, J., Choi, D., Zwickl, C., Morrison, N., Rashid, A., Hasan, A., Bao, W., Richard, A., Tong, W., Bushel, P. R., Brown, R., Bruno, M., Cunningham, M. L., Dix, D., Eastin, W., Frade, C., Garcia, A., Heinloth, A., Irwin, R., Madenspacher, J., Merrick, B. A., Papoian, T., Paules, R., Rocca-Serra, P., Sansone, A. S., Stevens, J., Tomer, K., Yang, C., & Waters, M. (2005). Chemical effects in biological systems–data dictionary (CEBS-DD): a compendium of terms for the capture and integration of biological study design description, conventional phenotypes, and ′omics data. Toxicological Sciences, 88, 585–601.
Jenkins, H., Hardy, N., Beckmann, M., Draper, J., Smith, A. R., Taylor, J., Fiehn, O., Goodacre, R., Bino, R. J., Hall, R., Kopka, J., Lane, G. A., Lange, B. M., Liu, J. R., Mendes, P., Nikolau, B. J., Oliver. S. G., Paton, N. W., Rhee, S., Roessner-Tunali, U., Saito, K., Smedsgaard, J., Sumner, L. W., Wang, T., Walsh, S., Wurtele, E. S., & Kell, D. B. (2004). A proposed framework for the description of plant metabolomics experiments and their results. Nature Biotechnology, 22, 1601–1606.
Jenkins, H., Johnson, H., Kular, B., Wang, T., & Hardy, N. (2005). Towards supportive data collection tools for plant metabolomics. Plant Physiology, 138, 67–77.
Lindon, J., Nicholson, J., Holmes, E., Keun, H., Craig, A., Pearce, J., Bruce, S., Hardy, N., Sansone, S., Antti, H., Jonsson, P., Daykin, C., Navarange, M., Beger, R., Verheij, E., Amberg, A., Baunsgaard, D., Cantor, G., Lehman-McKeeman, L., Earll, M., Wold, S., Johansson, E., Haselden, J., Kramer, K., Thomas, C., Lindberg, J., Schuppe-Koistinen, I., Wilson, I., Reily, M., Robertson, D., Senn, H., Krotzky, A., Kochhar, S., Powell, J., van der Ouderaa, F., Plumb, R., Schaefer, H., & Spraul, M. (2005). Summary recommendations for standardization and reporting of metabolic analyses. Nature Biotechnology, 23, 833–838.
Taylor, C. F. (2006). Minimum Reporting requirements for proteomics: A MIAPE Primer. Proteomics, 6, 39–44.
Trygg, J., Gullberg, J., Hohansson, A. I., Jonsson, P., & Moritz, T. (2006). Chemometrics in metabolomics—an introduction. In K. Saito, R. A. Dixon & L. Willmitzer (Eds.), Biotechnology in agriculture and forestry, Vol. 57. Plant metabolomics (pp. 117–128).
van der Werf, M. J. (2005). Towards replacing closed with open target selection approaches. Trends in Biotechnology, 23, 11–16.
Villas-Boas, S. G., Hojer-Pedersen, J., Akesson, M., Smedsgaard, J., & Nielsen, J. (2005). Global metabolite analysis of yeast: Evaluation of sample preparation protocols. Yeast, 22, 1155–1169.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License ( https://creativecommons.org/licenses/by-nc/2.0 ), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
van der Werf, M.J., Takors, R., Smedsgaard, J. et al. Standard reporting requirements for biological samples in metabolomics experiments: microbial and in vitro biology experiments. Metabolomics 3, 189–194 (2007). https://doi.org/10.1007/s11306-007-0080-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11306-007-0080-4