Abstract
Flower volatile organic compounds (VOCs) emitted from the receptive figs of Ficus (Moraceae) play important roles in maintaining the specificity of pollinator attraction and reproductive isolation between host species. The dominant components of VOCs are terpenoids, and while the genes involved in terpenoid biosynthesis are known, we know very little about the regulation of these genes and its relationship to pollinator attraction in Ficus. Thus, we selected pre-receptive (pre-VOC attractant-emitting) and receptive (VOC attractant-emitting) phase female flowers of a dioecious fig for transcriptome sequencing to investigate the differential activity of genes, including those related to terpenoid biosynthesis. We annoted 53,445 unigenes, of which 608 were found to be differently expressed in the two stages, with 242 unigenes upregulated and 366 unigenes downregulated in pre-receptive relative to receptive phase flowers. With respect to the production of VOCs, 80 unigenes encoded 34 enzyme-coding genes involved in terpenoid biosynthesis, including nearly all key enzymes of the MVA and MEP pathways forming the backbone of terpenoid biosynthesis. We designed pair primers for 16 of these 34 enzyme genes and validated 5 of them in 30 Ficus species representing 6 subgenera. In addition to the transcript sequences, we identified 35,558 putative microsatellite loci and designed primers for 11,468 of them. Our data and results will contribute to genetic research on terpene biosynthesis in figs and will aid in the understanding of the morphological and chemical changes that occur as the female flowers develop and produce pollinator-specific VOCs.

Similar content being viewed by others
References
Ahmed S, Dawson DA, Compton SG, Gilmartin PM (2007) Characterization of microsatellite loci in the African fig Ficus sycomorus L. (Moraceae). Mol Ecol Notes 7:1175–1177. doi:10.1111/j.1471-8286.2007.01822.x
Aubourg S, Lecharny A, Bohlmann J (2002) Genomic analysis of the terpenoid synthase (AtTPS) gene family of Arabidopsis thaliana. Mol Gen Genomics 267:730–745. doi:10.1007/s00438-002-0709-y
Barker NP (1985) Evidence of a volatile attractant in Ficus ingens (Moraceae). Bothalia 15:607–611. doi:10.4102/abc.v15i3/4.1854
Berg CC (2003) Flora Malesiana precursor for the treatment of Moraceae 1: the main subdivision of Ficus: the subgenera. Blumea 48:167–178. doi:10.3767/000651903X686132
Boutanaev AM, Moses T, Zi J, Nelson DR, Mugford ST, Peters RJ, Osbourn A (2014) Investigation of terpene diversification across multiple sequenced plant genomes. Proc Natl Acad Sci U S A 112:e81–e88. doi:10.1073/pnas.1419547112
Carretero-Paulet L, Cairo A, Botella-Pavia P, Besumbes O, Campos N, Boronat A, Rodriguez-Concepcion M (2006) Enhanced flux through the methylerythritol 4-phosphate pathway in Arabidopsis plants overexpressing deoxyxylulose 5-phosphate reductoisomerase. Plant Mol Biol 62:683–695. doi:10.1007/s11103-006-9051-9
Chen F (2003) Biosynthesis and emission of terpenoid volatiles from Arabidopsis flowers. Plant Cell 15:481–494. doi:10.1105/tpc
Chen C, Song Q (2008) Responses of the pollinating wasp Ceratosolen solmsi marchali to odor variation between two floral stages of Ficus hispida. J Chem Ecol 34:1536–1544. doi:10.1007/s10886-008-9558-4
Chen F, Tholl D, Bohlmann J, Pichersky E (2011) The family of terpene synthases in plants: a mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J 66:212–229. doi:10.1111/j.1365-313X.2011.04520.x
Cordoba E, Salmi M, Leon P (2009) Unravelling the regulatory mechanisms that modulate the MEP pathway in higher plants. J Exp Bot 60:2933–2943. doi:10.1093/jxb/erp190
Cordoba E, Porta H, Arroyo A, San Roman C, Medina L, Rodriguez-concepcion M, Leon P (2011) Functional characterization of the three genes encoding 1-deoxy-d-xylulose 5-phosphate synthase in maize. J Exp Bot 62:2023–2038. doi:10.1093/jxb/erq393
Crozier YC, Jia XC, Yao JY, Field AR, Cook JM, Crozier RH (2007) Microsatellite primers for Ficus racemosa and Ficus rubiginosa. Mol Ecol Notes 7:57–59. doi:10.1111/j.1471-8286.2006.01523.x
Cruaud A, Jabbour-Zahab R, Genson G et al (2010) Laying the foundations for a new classification of Agaonidae (Hymenoptera: Chalcidoidea), a multilocus phylogenetic approach. Cladistics 26:359–387. doi:10.1111/j.1096-0031.2009.00291.x
Cruaud A, Ronsted N, Chantarasuwan B et al (2012) An extreme case of plant-insect co-diversification: figs and fig-pollinating wasps. Syst Biol 61:1029–1047. doi:10.1093/sysbio/sys068
Cseke L, Dudareva N, Pichersky E (1998) Structure and evolution of linalool synthase. Mol Biol Evol 15:1491–1498
Degenhardt J, Köllner TG, Gershenzon J (2009) Monoterpene and sesquiterpene synthases and the origin of terpene skeletal diversity in plants. Phytochemistry 70:1621–1637. doi:10.1016/j.phytochem.2009.07.030
Dobson HEM, Bergström G, Groth I (1990) Differences in fragrance chemistry between flower parts of Rosa rugosa Thunb (Rosaceae). Israel J Bot 39:b143–b156. doi:10.1080/0021213X.1990.10677139
Dudareva N, Pichersky E, Gershenzon J (2004) Biochemistry of plant volatiles. Plant Physiol 135:1893–1902. doi:10.1104/pp. 104.049981
Dudareva N, Negre F, Negegowda DA, Orlova I (2006) Plant volatiles: recent advances and future perspectives. Crit Rev Plant Sci 25:417–440. doi:10.1080/07352680600899973
Dudareva N, Klempien A, Muhlemann JK, Kaplan I (2013) Biosynthesis, function and metabolic engineering of plant volatile organic compounds. New Phytol 198:16–32. doi:10.1111/nph.12145
Enfissi EM, Fraser PD, Lois LM, Boronat A, Schuch W, Bramley PM (2005) Metabolic engineering of the mevalonate and non-mevalonate isopentenyl diphosphate-forming pathways for the production of health-promoting isoprenoids in tomato. Plant Biotechnol J 3:17–27. doi:10.1111/j.1467-7652.2004.00091.x
Estévez JM, Cantero A, Reindl A, Reichler S, Leon P (2001) 1-Deoxy-d-xylulose-5-phosphate synthase, a limiting enzyme for plastidic isoprenoid biosynthesis in plants. J Biol Chem 276:22901–22909. doi:10.1007/978-1-4614-4063-5_2
Freiman ZE, Doron-Faigenboim A, Dasmohapatra R, Yablovitz Z, Flaishman MA (2014) High-throughput sequencing analysis of common fig (Ficus carica L.) transcriptome during fruit ripening. Tree Genet Genomes 10:923–935. doi:10.1007/s11295-014-0732-2
Garcia M, Bain A, Tzeng HY et al (2012) Portable microsatellite primers for Ficus (Moraceae). Am J Bot 99:e187–e192. doi:10.3732/ajb.1100485
Grabherr MG, Haas BJ, Yassour M et al (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29:644–652. doi:10.1038/nbt.1883
Grison L, Edwards AA, Hossaert-McKey M (1999) Interspecies variation in floral fragrances emitted by tropical Ficus species. Phytochemistry 52:1293–1299. doi:10.1016/S0031-9422(99)00411-2
Grison-Pigé L, Bessiére JM, Hossaert-McKey M (2002a) Specific attraction of fig-pollinating wasps: role of volatile compounds released by tropical figs. J Chem Ecol 28:283–295. doi:10.1023/A:1017930023741
Grison-Pigé L, Hossaert-McKey M, Greeff JM, Bessiére JM (2002b) Fig volatile compounds—a first comparative study. Phytochemistry 61:61–71. doi:10.1016/S0031-9422(02)00213-3
Guevara-Garcia A, San Roman C, Arroyo A et al (2005) Characterization of the Arabidopsis clb6 mutant illustrates the importance of posttranscriptional regulation of the methyl-d-erythritol 4-phosphate pathway. Plant Cell 17:628–643. doi:10.1105/tpc.104.028860
Guzman F, Kulcheski FR, Turchetto-Zolet AC, Margis R (2014) De novo assembly of Eugenia uniflora L. transcriptome and identification of genes from the terpenoid biosynthesis pathway. Plant Sci 229:238–245. doi:10.1016/j.plantsci.2014.10.003
Heer K, Machado CA, Himler AG, Herre EA, Kalko EK, Dick C (2012) Anonymous and EST-based microsatellite DNA markers that transfer broadly across the fig tree genus (Ficus, Moraceae). Am J Bot 99:e330–e333. doi:10.3732/ajb.1200032
Hossaert-Mckey M, Gibernau M, Frey JE (1994) Chemosensory attraction of fig wasps to substances produced by receptive figs. Entomol Exp Appl 70:185–191. doi:10.1111/j.1570-7458.1994.tb00746.x
Hossaert-McKey M, Soler C, Schatz B, Proffit M (2010) Floral scents: their roles in nursery pollination mutualism. Chemoecology 20:75–88. doi:10.1007/s00049-010-0043-5
Ikegami H, Habu T, Mori K, Nogata H, Hirata C, Hirashima K, Tashiro K, Kuhara S (2013) De novo sequencing and comparative analysis of expressed sequence tags from gynodioecious fig (Ficus carica L.) fruits: caprifig and common fig. Tree Genet Genomes 9:1075–1088. doi:10.1007/s11295-013-0622-z
Keeling CI, Bohlmann J (2006) Genes, enzymes and chemicals of terpenoid diversity in the constitutive and induced defence of conifers against insects and pathogens. New Phytol 179:657–675. doi:10.1111/j.1469-8137.2006.01716.x
Khadari B, Hochu I, Santoni S, Kjellberg F (2001) Identification and characterization of microsatellite loci in the common fig (Ficus carica L.) and representative species of genus Ficus. Mol Ecol Notes 1:191–193. doi:10.1046/j.1471-8278.2001.00072.x
Knudsen JT, Tollsten L, Bergström G (1993) Floral scents: a checklist of volatile compounds isolated by head-space techniques. Phytochemistry 33:253–280. doi:10.1016/0031-9422(93)
Knudsen JT, Tollsten L, Groth I et al (2004) Trends in floral scent chemistry in pollination syndromes: floral scent composition in hummingbird-pollinated taxa. Bot J Linn Soc 146:191–199. doi:10.1111/j.1095-8339.1993.tb00340.x
Koressaar T, Remm M (2007) Enhancements and modifications of primer design program Primer3. Bioinformatics 23:1289–1291. doi:10.1093/bioinformatics/btm091
Kumari S, Priya P, Misra G, Yadav G (2013) Structural and biochemical perspectives in plant isoprenoid biosynthesis. Phytochem Rev 12:255–291. doi:10.1007/s11101-013-9284-6
Lambert FR, Marshall AG (1991) Keystone characteristics of bird-dispersed Ficus in a Malaysian lowland rain forest. J Ecol 79:793–809. doi:10.2307/2260668
Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25. doi:10.1186/gb-2009-10-3-r25
Li B, Dewey CN (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. doi:10.1186/1471-2105-12-323
Moe AM, Weiblen GD (2011) Development and characterization of microsatellite loci in dioecious figs (Ficus, Moraceae). Am J Bot 98:e25–e27. doi:10.3732/ajb.1000412
Morris WL, Ducreux LJM, Hedden P, Millam S, Taylor MA (2006) Overexpression of a bacterial 1-deoxy-d-xylulose 5-phosphate synthase gene in potato tubers perturbs the isoprenoid metabolic network: implications for the control of the tuber life cycle. J Exp Bot 57:3007–3018. doi:10.1093/jxb/erl061
Nazareno AG, Santinelo Pereira RA, Feres JM, Mestriner MA, Alzate-Marin AL (2009) Transferability and characterization of microsatellite markers in two Neotropical Ficus species. Genet Mol Biol 32:568–571. doi:10.1590/S1415-47572009005000056
Oku H, Inafuku M, Takamine T, Nagamine M, Saitoh S, Fukuta M (2014) Temperature threshold of isoprene emission from tropical trees, Ficus virgata and Ficus septica. Chemosphere 95:268–273. doi:10.1016/j.chemosphere.2013.09.003
Phillips MA, Walter MH, Ralph SG et al (2007) Functional identification and differential expression of 1-deoxy-d-xylulose 5-phosphate synthase in induced terpenoid resin formation of Norway spruce (Picea abies). Plant Mol Biol 65:243–257. doi:10.1007/s11103-007-9212-5
Proffit M, Johnson SD (2009) Specificity of the signal emitted by figs to attract their pollinating wasps: comparison of volatile organic compounds emitted by receptive syconia of Ficus sur and F. sycomorus in Southern Africa. S Afr J Bot 75:771–777. doi:10.1016/j.sajb.2009.08.006
Raguso RA, Light DM, Pichersky E (1996) Electroantennogram responses of Hyles lineate (Sphingidoe: Lepidoptera) to volatile compounds from Clarkia breweri (Onagraceae) and other moth-pollinated flowers. J Chem Ecol 22:1735–1766. doi:10.1007/BF02028502
Ramsay H, Rieseberg LH, Ritland K (2009) The correlation of evolutionary rate with pathway position in plant terpenoid biosynthesis. Mol Biol Evol 26:1045–1053. doi:10.1093/molbev/msp021
Soler C, Hossaert-McKey M, Buatois B, Bessiere JM, Schatz B, Proffit M (2011) Georgraphic variation of floral scent in a highly specialized pollination mutualism. Phytochemistry 72:74–81. doi:10.1016/j.phytochem.2010.10.012
Storey JD, Tibshirani R (2003) Statistical significance for genomewide studies. PNAS 100:9440–9445. doi:10.1073/pnas.1530509100
Untergrasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG (2012) Primer3—new capabilities and interfaces. Nucleic Acids Res 40, e115. doi:10.1093/nar/gks596
Van Noort S, Ware AB, Compton SG (1989) Pollinator-specific volatile attractants released from figs of Ficus burtt-davyi. S Afr J Sci 85:323–324
Vignes H, Hossaert-McKey M, Beaune D, Fevre D, Anstett MC, Borges RM, Kjellberg F, Chevallier MH (2006) Development and characterization of microsatellite markers for a monoecious Ficus species, Ficus insipida, and crossspecies amplification among different sections of Ficus. Mol Ecol Notes 6:792–795. doi:10.1111/j.1471-8286.2006.01347.x
Wang Z, Gerstein M, Snyder M (2009) RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10:57–63. doi:10.1038/nrg2484
Wang L, Feng Z, Wang X, Wang X, Zhang X (2010) DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26:136–138. doi:10.1093/bioinformatics/btp612
Yu H, Compton SG (2012) Moving your sons to safety: galls containing male fig wasps expand into the centre of figs, away from enemies. PLoS ONE 7, e30833. doi:10.1371/journal.pone.0030833
Zavodna M, Arens P, Van Dijk PJ, Vosman B (2005) Development and characterization of microsatellite markers for two dioecious Ficus species. Mol Ecol Notes 5:355–357. doi:10.1111/j.1471-8286.2005.00924.x
Acknowledgments
The authors thank Prof. Zhang Mingyong, Dr. Xia Kuaifei, and Dr. Zeng Jiqing from South China Botanical Garden, the Chinese Academy of Sciences for their support with the transcriptome sequencing. We also thank three anonymous reviewers for extensive comments on this manuscript. This study was supported by the National Basic Research Program of China (973 Program) (2014CB954103), National Natural Science Foundation of China (31370409), Natural Science Foundation of Guangdong Province (S2013020012814), and the Applied Fundmental Research Foundation of Yunnan Province (2014GA003).
Conflict of interest
The authors declared that they have no competing interests.
Data archiving statement
Raw sequence reads are available from NCBI SRA (SRX951263 and SRX951272).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by C. Chen
This article is part of the Topical Collection on Genome Biology
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. A1
Overview of the transcriptome assembly of female flowers in male syconia of Ficus hirta. a. The size distribution of the transcripts. b. The size distribution of the unigenes. (GIF 1663 kb)
(GIF 1398 kb)
Fig. A2
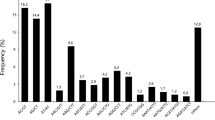
Characteristics of the homology search of Ficus hirta female flower unigenes. a. E-value distribution of the top BLASTx hits against the non-redundant (Nr) protein database for each unigene. The relative abundance of hits in each E-value range is represented as a percentage of the total number of hits. b. Species distribution of the top BLASTx hits in the NCBI nr database. (GIF 1637 kb)
Fig. A3
Distribution of most abundant gene ontology (GO) terms assigned to the Ficus hirta transcriptome. Proportions of annotated unigenes in the three GO categories 'Biological Process', 'Cellular Component' and 'Molecular Function' are reported for categories represented in at least the 0.1 % of the annotated unigenes. (PDF 6 kb)
Fig. A4
KOG function classification of the Ficus hirta transcriptome. In total, 8,376 unigenes were grouped into 25 classifications. (PDF 5 kb)
Fig. A5
KEGG function classification of the Ficus hirta transcriptome. In total, 3,990 unigenes are summarized into four main categories: A. cellular process; B. environmental information processing; C. genetic information processing; D. metabolism. (PDF 5 kb)
Fig A6
Functional categories of unigenes differentially expressed between the pre-receptive and receptive phase female flowers of Ficus hirta. Proportions of annotated unigenes in the three GO categories 'Biological Process' (BP), 'Cellular Component' (CC) and 'Molecular Function' (MF) are reported for categories represented in at least the 0.1 % of the annotated unigenes. (PDF 6 kb)
ESM 1
(PDF 69 kb)
Appendix 1
The putative SSR loci detected in the transcriptome sequence in Ficus hirta Vahl. Primer pair sequences are indicated for the subset of SSR loci for which Primer3 was able to design primers. For these loci, three pairs of primers were designed. (XLSX 3766 kb)
Rights and permissions
About this article
Cite this article
Yu, H., Nason, J.D., Zhang, L. et al. De novo transcriptome sequencing in Ficus hirta Vahl. (Moraceae) to investigate gene regulation involved in the biosynthesis of pollinator attracting volatiles. Tree Genetics & Genomes 11, 91 (2015). https://doi.org/10.1007/s11295-015-0916-4
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-015-0916-4