Abstract
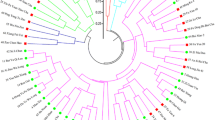
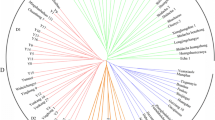
The improved clonal tea cultivars play a crucial role in the modern tea industry. In present study, we analyzed 128 elite clonal tea cultivars in China with 30 well-chosen simple sequence repeat (SSR) markers, aiming at (1) characterizing a set of DNA markers for unambiguously fingerprinting clonal tea cultivars and (2) confirming or identifying the parent-offspring (PO) relationships among them. The results showed that the markers are highly polymorphic among the tested cultivars with an average allele number of 10.4 per locus and an average polymorphic information content of 0.704. Robust fingerprinting power was demonstrated: It was possible to fully discriminate all 128 cultivars by a combination of four markers, and the overall possibility of finding two random individuals having the same genotypes across the 30 loci was estimated to be 4.8 × 10−33. Eight SSR loci were further recommended as a core marker set for fingerprinting the tea plant. Meanwhile, parentage analysis based on the fingerprint data revealed 47 pairs of putative PO relationships, among which 33 were in agreement with the known pedigree information, whereas the other 14 were newly identified in this study. The SSR markers and pedigree relationships reported here are valuable for tea cultivar identification and new breeding programs.



Similar content being viewed by others
References
Chen L, Zhou ZX, Yang YJ (2007) Genetic improvement and breeding of tea plant (Camellia sinensis) in China: from individual selection to hybridization and molecular breeding. Euphytica 154:239–248
Dakin EE, Avise JC (2004) Microsatellite null alleles in parentage analysis. Heredity 93:504–509
Duan YS, Jiang YH, Wang LY, Cheng H, Wang YH, Li XH (2011) Analysis of genetic diversity and relationship of tea cultivars and lines suitable for making green and black tea using SSR markers. Sci Agric Sin 44(1):99–109
Ellis JR, Burke JM (2007) EST-SSRs as a resource for population genetic analyses. Heredity 99:125–132
Fang W, Cheng H, Duan Y, Jiang X, Li X (2012) Genetic diversity and relationship of clonal tea (Camellia sinensis) cultivars in China as revealed by SSR markers. Plant Syst Evol 298:469–483
Fang WP, Meinhardt LW, Tan HW, Zhou L, Mischke S, Zhang D (2014) Varietal identification of tea (Camellia sinensis) using nanofluidic array of single nucleotide polymorphism (SNP) markers. Hortic Res 1:14035
Guichoux E, Lagache L, Wagner S, Léger P, Petit RJ (2011) Two highly validated multiplexes (12-plex and 8-plex) for species delimitation and parentage analysis in oaks (Quercus spp.). Mol Ecol Resour 11:578–585
Hayat K, Iqbal H, Malik U, Bilal U, Mushtaq S (2015) Tea and its consumption: benefits and risks. Crit Rev Food Sci Nutr 55(7):939–954
Jones A, Small CM, Paczolt KA, Ratterman NL (2010) A practical guide to methods of parentage analysis. Mol Ecol Resour 10:6–30
Kalinowski ST, Taper ML, Marshall TC (2007) Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol Ecol 16:1099–1106
Kaundun SS, Matsumoto S (2004) PCR-based amplicon length polymorphisms (ALPs) at microsatellite loci and indels from non-coding DNA regions of cloned genes as a means of authenticating commercial Japanese green teas. J Sci Food Agric 84:895–902
Lesser MR, Jackson ST (2013) Contributions of long-distance dispersal to population growth in colonising Pinus ponderosa populations. Ecol Lett 16:380–389
Li XH, Shi ZP, Liu CL, Luo JW, Shen CW, Gong ZH (2001) Parentage identification of filial generation tea plants from “Yunnan Daye” and “Rucheng Baimao” with RAPD method. J Tea Sci 21(2):99–102
Liang YR, Tanaka J, Takeda Y (2000) Parentage analysis of tea clone “Okumidori” with RAPD method. J Tea Sci 20(1):22–26
Liu K, Muse SV (2005) PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21:2128–2129
Liu Z, Zhao Y, Yang P, Yang C, Ling J, Yang Y (2014) Comparison of parents identification for tea variety based on SSR, SRAP and ISSR markers. J Tea Sci 34(6):617–624
Ma JQ, Zhou YH, Ma CL, Yao MZ, Jin JQ, Wang XC, Chen L (2010) Identification and characterization of 74 novel polymorphic EST-SSR markers in the tea plant, Camellia sinensis (Theaceae). Am J Bot 97:e153–e156
Ma JQ, Ma CL, Yao MZ, Jin JQ, Wang ZL, Wang XC, Chen L (2012) Microsatellite markers from tea plant expressed sequence tags (ESTs) and their applicability for cross-species/genera amplification and genetic mapping. Sci Hortic 134:167–175
Marshall TC, Slate J, Kruuk LEB, Pemberton JM (1998) Statistical confidence for likelihood-based paternity inference in natural populations. Mol Ecol 7:639–655
Nei M, Tajima FA, Tateno Y (1983) Accuracy of estimated phylogenetic trees from molecular data. J Mol Evol 19:153–170
Peakall R, Smouse PE (2006) Genalex 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics 28:2537–2539
Selkoe KA, Toonen RJ (2006) Microsatellites for ecologists: a practical guide to using and evaluating microsatellite markers. Ecol Lett 9:615–629
Sharangi AB (2009) Medicinal and therapeutic potentialities of tea (Camellia sinensis L.)—a review. Food Res Int 42:529–535
Summers K, Amos W (1997) Behavioral, ecological, and molecular genetic analyses of reproductive strategies in the Amazonian dart-poison frog, Dendrobates ventrimaculatus. Behav Ecol 8:260–267
Taberlet P, Luikart G (1999) Non-invasive genetic sampling and individual identification. Biol J Linn Soc 68:41–55
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Tan LQ, Wang LY, Wei K, Zhang CC, Wu LY, Qi GN, Cheng H, Zhang Q, Cui QM, Liang JB (2013) Floral transcriptome sequencing for SSR marker development and linkage map construction in the tea plant (Camellia sinensis). PLoS One 8(11):e81611
Taniguchi F, Furukawa K, Ohta-Metoku S, Yamaguchi N, Ujihara T, Kono I, Fukuoka H, Tanaka J (2012) Construction of a high density reference linkage map of tea (Camellia sinensis). Breed Sci 62:263–273
Taniguchi F, Kimura K, Saba T, Ogino A, Yamaguchi S, Tanaka J (2014) Worldwide core collections of tea (Camellia sinensis) based on SSR markers. Tree Genet Genomes 10(6):1555–1565
Tarroux E, DesRochers A, Tremblay F (2014) Molecular analysis of natural root grafting in jack pine (Pinus banksiana) trees: how does genetic proximity influence anastomosis occurrence? Tree Genet Genomes 10:667–677
Ujihara T, Ohta R, Hayashi N et al (2009) Identification of Japanese and Chinese green tea cultivars by using simple sequence repeat markers to encourage proper labeling. Biosci Biotechnol Biochem 73:15–20
Ujihara T, Taniguchi F, Tanaka J, Hayashi N (2011) Development of expressed sequence tag (EST)-based cleaved amplified polymorphic sequence (CAPS) markers of tea plant and their application to cultivar identification. J Agric Food Chem 59:1557–1564
van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) MICRO-CHACKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538
Wagner S, Gerber S, Petit RJ (2012) Two highly informative dinucleotide SSR multiplexes for the conifer Larix decidua (European larch). Mol Ecol Resour 12:717–725
Waits LP, Luikart G, Taberlet P (2001) Estimating the probability of identity among genotypes in natural populations: cautions and guidelines. Mol Ecol 10:249–256
Wang J, Santure AW (2009) Parentage and sibship inference from multilocus genotype data under polygamy. Genetics 181(4):1579–1594
Wang Y, Sun X, Li Y, Jiang H, Song W, Liu B, Zhang Y, Ma L, Yi B, Duan Z (2011) Parentage identification of newly hybrid tea cultivar Foxiang series revealed by inter-simple sequence repeat markers. Acta Agric Boreali-occidentalis Sin 20(7):149–154
Yang YJ, Liang YR (2014) Clonal tea cultivars in China. Shanghai Scientific and Technical Publishers, Shanghai
Yang YJ, Yang SJ, Yang YH, Zeng JM (2003) The breeding of an early sprouting and high quality new clone suitable for fine green tea. J China Tea 25(2):13–15
Yang CS, Wang X, Lu G, Picinich SC (2009) Cancer prevention by tea: animal studies, molecular mechanisms and human relevance. Nat Rev Cancer 9:429–439
Yang Y, Liu Z, Zhao Y, Liang GQ (2010) Construction of DNA fingerprints for tea cultivars originated from Hunan Province. J Tea Sci 30(5):367–373
Yao MZ, Ma CL, Qiao TT, Jin JQ, Chen L (2012) Diversity distribution and population structure of tea germplasms in China revealed by EST-SSR markers. Tree Genet Genomes 8:205–220
Zhang SG, Dong LJ, Yang Y, Liu Z, Wang X, Ning J (2009) Identification of male parents for “Yulv” and “Yusun” tea variety based on EST-SSR markers. J Tea Sci 29(6):430–435
Acknowledgments
This work was supported by the Twelfth Five-Year Project of Sichuan Province for Tea Plant Breeding, Science and Technology Department Project of Sichuan Province (2012-12CGZHZX0579) and The Earmarked Fund for Modern Agro-industry Technology Research System (nycytx-23).
Data archiving statement
We followed standard Tree Genetics and Genomes policy. All genotype data are provided in Online Resource 2.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Communicated by Y. Tsumura
This article is part of the Topical Collection on Germplasm Diversity
Electronic supplementary material
Below is the link to the electronic supplementary material.
Online Resource 1
(PDF 59 kb)
Online Resource 2
(XLSX 33 kb)
Rights and permissions
About this article
Cite this article
Tan, LQ., Peng, M., Xu, LY. et al. Fingerprinting 128 Chinese clonal tea cultivars using SSR markers provides new insights into their pedigree relationships. Tree Genetics & Genomes 11, 90 (2015). https://doi.org/10.1007/s11295-015-0914-6
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-015-0914-6