Abstract
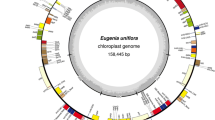
Since chloroplasts are maternally inherited and have unique features in evolution, their genome sequences have been broadly used in phylogenetic studies of plants. Here, we assembled the chloroplast genome sequence of cultivated pineapple (Ananas comosus (L.) Merr.) that is the most economically significant plant in the Bromeliaceae using next-generation sequencers. The genome length was 159,636 bp and included a pair of inverted repeats of 26,774 bp separated by a small single-copy region of 18,622 bp and a large single-copy region of 87,466 bp. The genome contained 113 unique genes (79 protein-coding, 4 rRNA, and 30 tRNA genes), 19 of which were duplicated in the inverted repeats, giving a total of 132 genes. We identified a total of 65 simple sequence repeats of >10 bp in length. Phylogenetic tree identified Ananas as a basal member of the Poales, closer to Musa (Musaceae, Zingiberales) than to species of the Poaceae. The genes, indels, and simple sequence repeats identified in this study will provide tools for use in evolutionary studies at both intra- and interspecific levels.






Similar content being viewed by others
References
Asano T, Tsudzuki T, Takahashi S, Shimada H, Kadowaki K (2004) Complete nucleotide sequence of the sugarcane (Saccharum officinarum) chloroplast genome: a comparative analysis of four monocot chloroplast genomes. DNA Res 11:93–99
Bausher MG, Singh ND, Mozoru J, Lee SB, Jansen RK, Daniell H (2006) The complete chloroplast genome sequence of Citrus sinensis (L.) Osbeck var. ‘Ridge Pineapple’: organization and phylogenetic relationships to other angiosperms. BMC Plant Biol 6:21
Bremer K (2000) Early cretaceous lineages of monocot flowering plants. Proc Natl Acad Sci U S A 97:4707–4711
Cato SA, Richardson TE (1996) Inter- and intraspecific polymorphism at chloroplast SSR loci and the inheritance of plastids in Pinus radiata D. Don. Theor Appl Genet 93:587–592
Chang CC, Lin HC, Lin IP, Chow TY, Chen HH, Chen WH, Cheng CH, Lin CY, Liu SM, Chaw SM (2006) The chloroplast genome of Phalaenopsis aphrodite (Orchidaceae): comparative analysis of evolutionary rate with that of grasses and its phylogenetic implications. Mol Biol Evol 23:279–291
Conant GC, Wolfe KH (2008) GenomeVx: simple web-based creation of editable circular chromosome maps. Bioinformatics 24:861–862
Coppens d’Eeckenbrugge G, Leal F (2002) Morphology, anatomy and taxonomy. In: Bartholomew DP, Paull RE, Rohrbach KG (eds) The pineapple: botany, production and uses. CAB International, Wallingford, pp 13–32
Coppens d’Eeckenbrugge G, Leal F, Duval MF (1997) Germplasm resources of pineapple. In: Janick J (ed) Horticultural reviews, vol 21. John Wiley and Sons, Hoboken, pp 133–175
Duval MF, Noyer JL, Perrier X, Coppens d’Eeckenbrugge G, Hamon P (2001) Molecular diversity in pineapple assessed by RFLP markers. Theor Appl Genet 102:83–90
Duval MF, Buso GS, Ferreira FR, Noyer JL, Coppens d’Eeckenbrugge G, Hamon P, Ferreira ME (2003) Relationships in Ananas and other related genera using chloroplast DNA restriction site variation. Genome 46:990–1004
Gilles A, Meglécz E, Pech N, Ferreira S, Malausa T, Martin JF (2011) Accuracy and quality assessment of 454 GS-FLX Titanium pyrosequencing. BMC Genomics 12:245
Goremykin VV, Holland B, Hirsch-Ernst KI, Hellwig FH (2005) Analysis of Acorus calamus chloroplast genome and its phylogenetic implications. Mol Biol Evol 22:1813–1822
Guisinger MM, Chumley TW, Kuehl JV, Boore JL, Jansen RK (2010) Implications of the plastid genome sequence of Typha (Typhaceae, Poales) for understanding genome evolution in Poaceae. J Mol Evol 70:149–166
Hamdan N, Samad AA, Hidyat T, Salleh FM (2013) Phylogenetic analysis of eight Malaysian pineapple cultivars using a chloroplast marker (rcbL gene). J Teknol 64:29–33
Hepton A (2002) Culture system. In: Bartholomew DP, Paull RE, Rohrbach KG (eds) The pineapple: botany, production and uses. CAB International, Wallingford, pp 109–142
Hiratsuka J, Shimada H, Whittier R, Ishibashi T, Sakamoto M, Mori M, Kondo C, Honji Y, Sun CR, Meng BY et al (1989) The complete sequence of the rice (Oryza sativa) chloroplast genome: intermolecular recombination between distinct tRNA genes accounts for a major plastid DNA inversion during the evolution of the cereals. Mol Gen Genet 217:185–194
Ishii T, Xu Y, McCouch SR (2001) Nuclear and chloroplast microsatellite variation in a genome species of rice. Genome 44:658–666
Jansen RK, Kaittanis C, Saski C, Lee SB, Tomkins J, Alverson AJ, Daniell H (2006) Phylogenetic analyses of Vitis (Vitaceae) based on complete chloroplast genome sequences: effects of taxon sampling and phylogenetic methods on resolving relationships among rosids. BMC Evol Biol 6:32
Jansen RK, Cai Z, Raubeson LA, Daniell H, dePamphilis CW, Leebens-Mack J, Muller KF, Guisinger-Bellian M, Haberle RC, Hansen AK et al (2007) Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc Natl Acad Sci U S A 104:19369–19374
Katayama H, Ogihara Y (1996) Phylogenetic affinities of the grasses to other monocots as revealed by molecular analysis of chloroplast DNA. Curr Genet 29:572–581
Kim KJ, Lee HL (2004) Complete chloroplast genome sequences from Korean ginseng (Panax schinseng Nees) and comparative analysis of sequence evolution among 17 vascular plants. DNA Res 11:247–261
Kim JS, Jung JD, Lee JA, Park HW, Oh KH, Jeong WJ, Choi DW, Liu JR, Cho KY (2006) Complete sequence and organization of the cucumber (Cucumis sativus L. cv. Baekmibaekdadagi) chloroplast genome. Plant Cell Rep 25:334–340
Leebens-Mack J, Raubeson LA, Cui L, Kuehl J, Fourcade M, Chumley T, Boore JL, Jansen RK, dePamphilis CW (2005) Identifying the basal angiosperms in chloroplast genome phylogenies: sampling one’s way out of the Felsenstein zone. Mol Biol Evol 22:1948–1963
Maier RM, Neckermann K, Igloi GL, Kossel H (1995) Complete sequence of the maize chloroplast genome: gene content, hotspots of divergence and fine tuning of genetic information by transcript editing. J Mol Biol 251:614–628
Malézieux E, Côte F, Bartholomew DP (2002) Crop environment, plant growth and physiology. In: Bartholomew DP, Paull RE, Rohrbach KG (eds) The pineapple: botany, production and uses. CAB International, Wallingford, pp 69–108
Martin G, Baurens FC, Cardi C, Aury JM, D’Hont A (2013) The complete chloroplast genome of banana (Musa acuminata, Zingiberales): insight into plastid monocotyledon evolution. PLoS One 8:e67350
Naito K, Kaga A, Tomooka N, Kawase M (2013) De novo assembly of the complete organelle genome sequences of azuki bean (Vigna angularis) using next-generation sequencers. Breed Sci 63:176–182
Ogihara Y, Isono K, Kojima T, Endo A, Hanaoka M, Shiina T, Terachi T, Utsugi S, Murata M, Mori N et al (2000) Chinese spring wheat (Triticum aestivum L.) chloroplast genome: complete sequence and contig clones. Plant Mol Biol Rep 18:243–253
Paz EY, Gil K, Rebolledo L, Rebolledo A, Uriza D, Martinez O, Isidron M, Simpson J (2005) AFLP characterization of the Mexican pineapple germplasm collection. J Am Soc Hort Sci 130:575–579
Paz EY, Gil K, Rebolledo L, Rebolledo A, Uriza D, Martinez O, Isidron M, Diaz L, Lorenzo JC, Simpson J (2012) Genetic diversity of Cuban pineapple germplasm assessed by AFLP markers. Crop Breed Appl Biotech 12:104–110
Powell W, Morgante M, McDevitt R, Vendramin GG, Rafalski JA (1995) Polymorphic simple sequence repeat regions in chloroplast genomes: applications to the population genetics of pines. Proc Natl Acad Sci U S A 92:7759–7763
Raubeson LA, Peery R, Chumley T, Dziubek C, Fourcade HM, Boore JL, Jansen RK (2007) Comparative chloroplast genomics: analyses including new sequences from the angiosperms Nuphar advena and Ranunculus macranthus. BMC Genomics 8:174
Ravi V, Khurana JP, Tyagi AK, Khurana P (2006) The chloroplast genome of mulberry: complete nucleotide sequence, gene organization and comparative analysis. Tree Genet Genomes 3:49–59
Rohrbach KG, Leal F, Coppens d’Eeckenbrugge G (2002) History, distribution and world production. In: Bartholomew DP, Paull RE, Rohrbach KG (eds) The pineapple: botany, production and uses. CAB International, Wallingford, pp 1–12
Ruhlman T, Lee SB, Jansen RK, Hostetler JB, Tallon LJ, Town CD, Daniell D (2006) Complete plastid genome sequence of Daucus carota: implications for biotechnology and phylogeny of angiosperms. BMC Genomics 7:224
Schwartz S, Zhang Z, Frazer K, Smit A, Riemer C, Bouck J, Gibbs R, Hardison R, Miller W (2000) PipMaker: a web server for aligning two genomic DNA sequences. Genome Res 10:577–586
Stamatakis A (2014) RAxML Version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313
Tangphatsornruang S, Sangsrakru D, Chanprasert J, Uthaipaisanwong P, Yoocha T, Jomchai N, Tragoonrung S (2010) The chloroplast genome sequence of mungbean (Vigna radiata) determined by high-throughput pyrosequencing: structural organization and phylogenetic relationships. DNA Res 17:11–22
Terakami S, Matsumura Y, Kurita K, Kanamori H, Katayose Y, Yamamoto T, Katayama H (2012) Complete sequence of the chloroplast genome from pear (Pyrus pyrifolia): genome structure and comparative analysis. Tree Genet Genomes 8:841–854
West-Eberhard MJ, Smith JA, Winter K (2011) Photosynthesis, reorganized. Science 332:311–312
Wyman SK, Boore JL, Jansen RK (2004) Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20:3252–3255
Acknowledgments
This study was partially supported by an Okinawa special promotion grant.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by J. L. Wegrzyn
Data Archiving Statement
Ananas comosus chloroplast DNA complete sequence is submitted to DDBJ; the accession number of the sequence is AP014632. All the sequence data (.fastq files) were deposited in the DDBJ Sequence Read Archive (accession: DRA002476).
This article is part of the Topical Collection on Genome Biology
Rights and permissions
About this article
Cite this article
Nashima, K., Terakami, S., Nishitani, C. et al. Complete chloroplast genome sequence of pineapple (Ananas comosus). Tree Genetics & Genomes 11, 60 (2015). https://doi.org/10.1007/s11295-015-0892-8
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-015-0892-8