Abstract
Islands isolated by oceans that act as a geographical barrier for plant migration often possess high species endemism and have been deemed as a natural laboratory for studying species divergence. Fatsia Decne. & Planch. (Araliaceae), with three species, is one of the few plant genera absent in continents while exclusively spanning continental and oceanic islands. The nuclear ribosomal internal transcribed spacer (nrITS) phylogeny uncovered a pattern with reciprocal monophyly of Fatsia oligocarpella Koidz. (Bonin) and Fatsia polycarpa Hayata (Taiwan) vs. paraphyly of Fatsia japonica (Thunb.) Decne. & Planch. (Japan and Ryukyus), suggesting ancestry of the species in Japan and a likely temperate origin; whereas, lack of monophyly of all three allopatrically distributed species at chloroplast DNA (cpDNA) trnL–trnF spacer likely resulted from lineage sorting. In spite of the limited habitats for F. oligocarpella, unexpectedly high genetic variations in this species of oceanic islands were likely attributable to multiple colonizations and recurrent gene introgression. Biogeographical analyses suggested that Fatsia likely diverged via southward colonization in Bonin Islands and Taiwan during the late Pliocene to Pleistocene. Besides, Fatsia species with an allopatric distribution provide a perfect model for testing speciation modes of insular endemics. Nonzero gene flow between species was detected based on MIGRATE and STRUCTURE analyses of DNA sequences and microsatellite fingerprints, suggesting that allopatric speciation is less likely.





Similar content being viewed by others
References
Asami S (1970) Topography and geology in the Bonin Islands. In: Tsuyama T, Asami S (eds) The nature of the Bonin Islands. Hirokawa Shoten, Tokyo, pp 91–108
Beerli P, Felsenstein J (2001) Maximum likelihood estimation of a migration matrix and effective population sizes in n subpopulations by using a coalescent approach. Proc Natl Acad Sci U S A 98:4563–4568
Chen CH (2000) The phylogenetic study of Gentiana sect. Chondrophyllae Bunge. Dissertation, Department of Life Science, National Taiwan Normal University, Taipei, Taiwan
Chiang TY, Schaal BA (2006) Phylogeography of plants in Taiwan and the Ryukyu Archipelago. Taxon 55:31–41
Chiang YC, Hung KH, Schaal BA, Ge XJ, Hsu TW, Chiang TY (2006) Contrasting phylogeographical patterns between mainland and island taxa of the Pinus luchensis complex. Mol Ecol 15:765–779
Chiang YC, Hung KH, Moore SJ, Ge XJ, Huang S, Hsu TW, Schaal BA, Chiang TY (2009) Paraphyly of organelle DNAs in Cycas sect. Asiorientales due to ancient ancestral polymorphisms. BMC Evol Biol 9:161
Conti E, Soltis DE, Hardig TM, Schneider J (1999) Phylogenetic relationships of the silver saxifrages (Saxifraga, sect. Ligulatae haworth): implications for the evolution of substrate specificity, life histories, and biogeography. Mol Phylogenet Evol 13:536–555
Cox CB, Moore PD (2010) Biogeography: an ecological and evolutionary approach. Blackwell Science Ltd, Oxford
Crandall KA, Templeton AR (1993) Empirical tests of some predictions from coalescence theory with applications to intraspecific phylogeny reconstruction. Genetics 134:959–969
Crawford DJ, Stuessy TF, Cosner MB, Haines M, Silva OM, Baeza M (1992) Evolution of the genus Dendroseris (Asteraceae: Lactuceae) in the Juan Fernandez Islands: evidence from chloroplast and ribosomal DNA. Syst Bot 17:676–682
Darwin CR (1859) On the origin of species. John Murray, London
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Drummond AJ, Ho SY, Phillips MJ, Rambaut A (2006) Relaxed phylogenetics and dating with confidence. PLoS Biol 4:e88
Dupanloup I, Schneider S, Excoffier L (2002) A simulated annealing approach to define the genetic and structure of populations. Mol Ecol 11:2571–2581
Earl DA, vonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Smouse PE (1994) Using allele frequencies and geographic subdivision to reconstruct gene trees within a species: molecular variance parsimony. Genetics 136:343–359
Excoffier L, Laval G, Schneider S (2007) Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform Online 1:47–50
Felsenstein J (2007) PHYLIP (Phylogeny Inference Package), version 3.67. Department of Genetics, University of Washington, Seattle
Fernández-Mazuecos M, Vargas P (2011) Genetically Depauperate in the Continent but Rich in Oceanic Islands: Cistus monspeliensis (Cistaceae) in the Canary Islands. PLoS ONE 6(2):e17172
Francisco-Ortega J, Santos-Guerra A, Kim SC, Crawford DJ (2000) Plant genetic diversity in the Canary Islands: a conservation perspective. Am J Bot 87:909–919
Givnish TJ (1998) Adaptive plant evolution on islands: classical patterns, molecular data, new insights. In: Grant PR (ed) Evolution on islands. Oxford University Press, New York, pp 284–301
Graur D, Li WH (2000) Fundamentals of molecular evolution. Sinauer Associates, Sunderland
Hiramatsu M, Ii K, Okubo H, Huang KL, Huang CW (2001) Biogeography and origin of Lilium longiflorum and L. formosanum (Liliaceae) endemic to the Ryukyu Archipelago and Taiwan as determined by allozyme diversity. Am J Bot 88:1230–1239
Huang S, Chiang YC, Schaal BA, Chou CH, Chiang TY (2001) Organelle DNA phylogeography of Cycas taitungensis, a relict species in Taiwan. Mol Ecol 10:2669–2681
Huang CC, Hung KH, Hsu TW, Wang KH, Lin CY, Chiang TY (2008) Isolation and characterization of 11 polymorphic microsatellite loci from Fatsia polycarpa (Araliaceae), an element of evergreen forests in Taiwan. Conserv Genet 9:1333–1335
Ito M (1998) Origin and evolution of endemic plants of the Bonin (Ogasawara) Islands. Res Popul Ecol 40:205–212
Ito M, On M (1990) Allozyme diversity and the evolution of Crepidiastrum (Compositae) on the Bonin (Ogasawara) Islands. Bot Mag Tokyo 103:449–459
Jukes TH, Cantor CR (1969) Evolution of protein molecules. In: Munro HN (ed) Mammalian protein metabolism. Academic Press, New York, pp 21–132
Kato Y, Yagi T (2004) Biogeography of the subspecies of Parides (Byasa) alcinous (Lepidoptera: Papilionidae) based on a phylogenetic analysis of mitochondrial ND5 sequences. Syst Entomol 29:1–9
Kay KM, Whittall JB, Hodges SA (2006) A survey of nuclear ribosomal internal transcribed spacer substitution rates across angiosperms: an approximate molecular clock with life history effects. BMC Evol Biol 6:36
Kimura M (1996) Quaternary palaeogeography of the Ryukyu Arc. J Geogr 105:259–285
Kizaki K, Oshiro I (1980) The origin of the Ryukyu Islands. In: Kizaki K (ed) Natural history of the Ryukyus. Tsukiji-Shokan, Tokyo, pp 8–37
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948
Li J, Zhang D, Donoghue MJ (2003) Phylogeny and biogeography of Chamaecyparis (Cupressaceae) inferred from DNA sequences of the nuclear ribosomal ITS region. Rhodora 105:106–117
Librado P, Rozas J (2009) DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452
Maki M, Morita H, Oiki S, Takahashi H (1999) The effect of geographic range and dichogamy on genetic variability and population genetic structure in Tricyrtis section Flavae (Liliaceae). Am J Bot 86:287–292
Maki M, Yamashiro T, Matsumura S (2003) High levels of genetic diversity in island populations of the island endemic Suzukia luchuensis (Labiatae). Heredity 91:300–306
Nakamura K, Denda T, Kokubugata G, Suwa R, Yang TYA, Peng CI, Yokota M (2010) Phylogeography of Ophiorrhiza japonica (Rubiaceae) in continental islands, the Ryukyu Archipelago, Japan. J Biogeogr 37:1907–1918
Nei M, Tajima F (1983) Maximum likelihood estimation of the number of nucleotide substitutions from restriction sites data. Genetics 105:207–217
Osada N (2004) Inferring the mode of speciation from genomic data: a study of the great apes. Genetics 169:259–264
Ota H (1998) Geographic patterns of endemism and speciation in amphibians and reptiles of the Ryukyu Archipelago, Japan, with special reference to their paleogeographical implications. Res Popul Ecol 40:189–204
Percy DM, Garver AM, Wagner WL, James HF, Cunningham CW, Miller SE, Fleischer RC (2008) Progressive island colonization and ancient origin of Hawaiian Metrosideros (Myrtaceae). Proc R Soc B Biol Sci 275:1479–1490
Plunkett GM, Wen J, Lowry PP (2004) Infrafamilial classifications and characters in Araliaceae: insights from the phylogenetic analysis of nuclear (ITS) and plastid (trnL–trnF) sequence data. Plant Syst Evol 245:1–39
Posada D, Crandall KA (1998) MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817–818
Pritchard J, Wen W (2003) Documentation for STRUCTURE Software, version 3.2. http://pritch.bsd.uchicago.edu/structure.html
Pritchard J, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Rambaut A, Drummond AJ (2004) Tracer—MCMC trace analysis tool. University of Oxford, Oxford
Sánchez JL, Domina G, Caujapé-Castells J (2005) Genetic differentiation of three species of Matthiola (Brassicaceae) in the Sicilian insular system. Plant Syst Evol 253:81–93
Sarno RJ, Franklin WL, O’Brien SJ, Johnson WE (2001) Patterns of mtDNA and microsatellite variation in an island and mainland population of guanacos in southern Chile. Anim Conserv 4:93–101
Seo A, Watanabe M, Hotta M, Murakami N (2004) Geographical patterns of allozyme variation in Angelica japonica (Umbelliferae) and Farfugium japonicum (Compositae) on the Ryukyu Islands, Japan. Acta Phytotax Geobot 55:29–44
Setoguchi H, Mitsui Y, Ikeda H, Nomura N, Tamura A (2011) Genetic structure of the critically endangered plant Tricyrtis ishiiana (Convallariaceae) in relict populations of Japan. Conserv Genet 12:491–501
Sibuet JC, Hsu SK (2004) How was Taiwan created? Tectonophysics 379:159–181
Soejima A, Nagamasu H, Ito M, Ono M (1994) Allozyme diversity and the evolution of Symplocos (Symplocaceae) on the Bonin (Ogasawara) Islands. J Plant Res 107:221–227
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595
Talavera MA, Navarro-Sampedro L, Ortiz PL, Arista M (2013) Phylogeography and seed dispersal in islands: the case of Rumex bucephalophorus subsp. canariensis (Polygonaceae). Ann Bot 111:249–260
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Vellend M (2003) Island biogeography of genes and species. Am Nat 162:358–365
Vita-Finzi C (2000) Deformation and seismicity of Taiwan. Proc Natl Acad Sci U S A 97:11176–11180
Wang WK, Ho CW, Hung KH, Wang KH, Huang CC, Osada N, Chiang TY (2010) Multi-locus analysis of genetic divergence between outcrossing Arabidopsis species: evidence of genome-wide admixture. New Phytol 188:488–500
Wolfe KH, Li WH, Sharp PM (1987) Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc Natl Acad Sci U S A 84:9054–9058
Wright S (1940) Breeding structure of populations in relation to speciation. Am Nat 74:232–248
Acknowledgments
This work was supported by the National Science Council of Taiwan [grant number NSC 95-2621-B-006-003-MY3].
Data Archiving Statement
All sequences of haplotypes were deposited with the European Molecular Biology Laboratory (EMBL) Nucleotide Sequence Database. The accession numbers are HG317014–HG317047 for cpDNA and HG317048–HG317098 for nrITS.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Communicated by W. Ratnam
T.-Y. Chiang, S.-F. Chen, H. Kato, and C.-C. Hwang equally contributed to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig. 1S
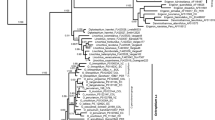
Maximum likelihood tree of Fatsia species rooted at outgroup sequences based on combined dataset. The bootstrap values are indicated at nodes (GIF 78 kb)
Supplementary Table 1
(DOC 56 kb)
Supplementary Table 2
(DOC 52 kb)
Supplementary Table 3
(DOC 61 kb)
Rights and permissions
About this article
Cite this article
Chiang, TY., Chen, SF., Kato, H. et al. Temperate origin and diversification via southward colonization in Fatsia (Araliaceae), an insular endemic genus of the West Pacific Rim. Tree Genetics & Genomes 10, 1317–1330 (2014). https://doi.org/10.1007/s11295-014-0763-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11295-014-0763-8