Abstract
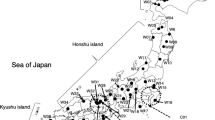
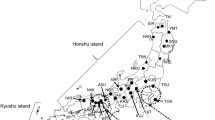
The Japanese chestnut (Castanea crenata Siebold et Zucc.) is naturally distributed throughout Japan and is cultivated for its fruit (nuts) throughout the country. Many native cultivars have cultivation records going back more than 100 years. Researchers have speculated that native cultivars that originated in the Tanba region, the most important region of cultivation, were spread throughout Japan. To clarify the breeding history and spreading pattern of Japanese chestnut cultivars, we estimated the population structure of a set of 60 native chestnut cultivars via hierarchical clustering and Bayesian model-based clustering. Both analyses gave similar results. The cultivars were divided into two main clusters: one with cultivars from the Tanba region, the other with cultivars from other areas of Japan. However, there were some exceptions to this pattern, suggesting that propagation of clones and seeds by humans was a part of the spreading process. Additionally, parent–offspring relationships were estimated from the data obtained for 175 simple sequence repeat markers. Out of the 60 genotypes, nine putative parent–offspring pairs and eight putative parent–offspring trios were identified. These results suggest that native cultivars are likely to have been selected from crosses of older native cultivars. In particular, some native cultivars from outside the Tanba region had parent–offspring relationships with cultivars from the Tanba region. This result suggests that cultivars from outside the Tanba region had been crossed with cultivars from the Tanba region and then selected as a means of introducing favorable traits from the Tanba cultivars.



Similar content being viewed by others
References
Boccacci P, Aramini M, Valentini N, Bacchetta L, Rovira M, Drogoudi P, Silva AP, Solar A, Calizzano F, Erdogan V, Cristofori V, Ciarmiello LF, Contessa C, Ferreira JJ, Marra FP, Botta R (2013) Molecular and morphological diversity of on-farm hazelnut (Corylus avellana L.) landraces from southern Europe and their role in the origin and diffusion of cultivated germplasm. Tree Genet Genomes 9:1465–1480
Boursiquot JM, Lacombe T, Laucou V, Julliard S, Perrin FX, Lanier N, Legrand D, Meredith C, This P (2009) Parentage of Merlot and related winegrape cultivars of southwestern France: discovery of the missing link. Aust J Grape Wine Res 15:144–155
Bowcock AM, Ruiz-Linares A, Tomfohrde J, Minch E, Kidd JR, Cavalli-Sforza LL (1994) High-resolution of human evolutionary trees with polymorphic microsatellites. Nature 368:455–457
Bowers J, Boursiquot JM, This P, Chu K, Johansson H, Meredith C (1999) Historical genetics: the parentage of Chardonnay, Gamay, and other wine grapes of northeastern France. Science 285:1562–1565
Cipriani G, Marrazzo MT, Peterlunger E (2010) Molecular characterization of the autochthonous grape cultivars of the region Friuli Venezia Giulia—North-Eastern Italy. Vitis 49:29–38
Comes HP, Kadereit JW (1998) The effect of quaternary climatic changes on plant distribution and evolution. Trends Plant Sci 3:432–438
Crespan M, Calo A, Giannetto S, Sparacio A, Storchi P, Costacurta A (2008) ‘Sangiovese’ and ‘Garganega’ are two key varieties of the Italian grapevine assortment evolution. Vitis 47:97–104
Di Vecchi-Staraz M, Bandinelli R, Boselli M, This P, Boursiquot JM, Laucou V, Lacombe T, Vares D (2007) Genetic structuring and parentage analysis for evolutionary studies in grapevine: kin group and origin of the cultivar Sangiovese revealed. J Am Soc Hort Sci 132:514–524
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Fujii H, Yamashita H, Hosaka F, Terakami S, Yamamoto T (2010) Development of a software to presume the parent-child relationship using the result of DNA marker typing. Hort Res (JAPAN) 9(1):34
Hewitt G (2000) The genetic legacy of the Quaternary ice ages. Nature 405:907–913
Inoue E, Ning L, Hara H, Ruan SA, Anzai H (2009) Development of simple sequence repeat markers in Chinese chestnut and their characterization in diverse chestnut cultivars. J Am Soc Hort Sci 134:610–617
Isaki M (1978) Theory and actual condition of chestnut cultivation. Hakuyusya, Tokyo (In Japanese)
Lacombe T, Boursiquot JM, Laucou V, Di Vecchi-Staraz M, Peros JP, This P (2013) Large-scale parentage analysis in an extended set of grapevine cultivars (Vitis vinifera L.). Theor Appl Genet 126:401–414
Mattioni C, Cherubini M, Micheli E, Villani F, Bucci G (2008) Role of domestication in shaping Castanea sativa genetic variation in Europe. Tree Genet Genomes 4:563–574
Moriya S, Inoue K, Otake A, Shiga M, Mabuchi M (1989) Decline of the chestnut gall wasp population, Dryocosmus kuriphilus Yasumatsu (Hymenoptera, Cynipidae) after the establishment of Torymus sinensis Kamijo (Hymenoptera, Torymidae). Appl Entomol Zool 24:231–233
Nishio S, Takada N, Yamamoto T, Terakami S, Hayashi T, Sawamura Y, Saito T (2013) Mapping and pedigree analysis of the gene that controls the easy peel pellicle trait in Japanese chestnut (Castanea crenata Sieb. et Zucc.). Tree Genet Genomes 9:723–730
Nishio S, Yamamoto T, Terakami S, Sawamura Y, Takada N, Nishitani C, Saito T (2011a) Novel genomic and EST-derived SSR markers in Japanese chestnuts. Sci Hortic 130:838–846
Nishio S, Yamamoto T, Terakami S, Sawamura Y, Takada N, Saito T (2011b) Genetic diversity of Japanese chestnut cultivars assessed by SSR markers. Breed Sci 61:109–120
Osaki M (1949) Fruit horticulture consultation. Asakurasyoten, Tokyo (In Japanese)
Pereira-Lorenzo S, Costa RML, Ramos-Cabrer AM, Ciordia-Ara M, Ribeiro CAM, Borges O, Barreneche T (2011) Chestnut cultivar diversification process in the Iberian Peninsula, Canary Islands, and Azores. Genome 54:301–315
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Sawamura Y (2006) Chestnut. In: Jpn. Soc. Hort. Sci. (ed) Horticulture in Japan 2006. Shoukadoh, Kyoto pp 82–85
Shimura I (1984) Cultivation in Japan. In: Rural Culture Association (ed) Nogyo-gijyutsu-taikei: Kajyu-hen Vol. 5. Rural Culture Association, Tokyo, pp 7–8 (in Japanese).
Shimura I (2003) Chestnut breeding history. Agric and Hortic 58:30–32 (in Japanese)
Suzuki R, Shimodaira H (2006) Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 22:1540–1542
Takada N, Nishio S, Yamada M, Sawamura Y, Sato A, Hirabayashi T, Saito T (2012) Inheritance of the easy-peeling pellicle trait of Japanese chestnut cultivar Porotan. HortSci 47:845–847
Tanaka T, Yamamoto T, Suzuki M (2005) Genetic diversity of Castanea crenata in northern Japan assessed by SSR markers. Breed Sci 55:271–277
Tanaka Y (1933) Cultivation of chestnut. Meibundou, Tokyo (In Japanese)
This P, Lacombe T, Thomas MR (2006) Historical origins and genetic diversity of wine grapes. Trends Genet 22:511–519
Tsukada M (1988) Japan. In: Huntley B, Webb T III (eds) Vegetation history. Kluwer, London, pp 459–518
Ward JH (1963) Hierarchical grouping to optimize an objective function. J Am Stat Assoc 58:236–244
Yagioka S (1915) Cultivation of chestnut. Dainihonnougyosyoreikai, Tokyo (In Japanese)
Yamamoto T, Terakami S, Shigeta T, Nishitani C, Nishio S, Saito T, Fuji H (2012) DNA profiling of fresh and processed nuts in Japanese chestnut. DNA Polymorphism 4:29–38 (In Japanese)
Acknowledgments
We are deeply indebted to all the people involved in the Japanese chestnut breeding program at the NARO Institute of Fruit Tree Science, Japan.
Data Archiving Statement
JP (Genebank) accession numbers of cultivars, SSR markers, and SSR genotype data used in this study are available as supplemental text files that can be downloaded with the manuscript. All SSR sequences have been deposited in the DNA Data Bank of Japan (DDBJ).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by A. Kremer
Rights and permissions
About this article
Cite this article
Nishio, S., Iketani, H., Fujii, H. et al. Use of population structure and parentage analyses to elucidate the spread of native cultivars of Japanese chestnut. Tree Genetics & Genomes 10, 1171–1180 (2014). https://doi.org/10.1007/s11295-014-0751-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11295-014-0751-z