Abstract
Although rabies virus is widely distributed in the world, and has been the subject of extensive investigations with the objective of its ultimate prevention, control, and management, there is much less knowledge of the characteristics, distribution, and infectivity of other lyssaviruses. Since bats are known animal vectors for all but one of the known lyssavirus genotypes, we have performed an extensive survey of bats in the Guangxi Province to provide information on lyssavirus distribution in southern China. The lyssavirus nucleoprotein gene was detected in brains of 2.86 % of 2,969 bats. Nucleotide sequence homologies among isolates were 86.9–99.6 %, but only 70.0–85.0 % for lyssaviruses in GenBank. These infected bats were detected from a wide area, essentially forming a band running from the south-west to the north-east of Guangxi, and it appears that infection by new lyssaviruses is widespread in this region.
Similar content being viewed by others
Introduction
In China incidents of rabies virus infection have grown dramatically during the 21st century [1], and this virus is currently responsible for more deaths than any other infectious agents. Thus, control of rabies is now one of the most important public health concerns in the country. While dog bites are undoubtedly the current principal mechanism for current human rabies infection, wild animal populations represent a potentially large reservoir for rabies and other lyssaviruses. Apart from rabies, the other lyssaviruses have not been studied sufficiently for their potential host range and pathogenicity to be evaluated. However, each of lyssavirus genotypes 2–7 is currently limited to a particular geographical area; Lagos bat virus (LBV) [2], Mokola virus (MOKV) [3], and Duvenhage virus (DUVV) [4] are found in Africa, the European bat virus-1 (EBLV-1) [5] and European bat virus-2 (EBLV-2) [6] have been isolated from European animals, and the Australian bat lyssavirus (ABLV) [7] has been detected on the Australian continent. The rabies virus is the only lyssavirus that has been detected in the Americas.
At present there is little information about bat lyssaviruses in Asia, despite the fact that India and China, homes to approximately 1/3 of the World’s people, are the two countries with the highest incidence of rabies [8]. Although antibodies to the rabies virus have been detected in sera of bats from southern China [9], and no infections with lyssavirus Genotypes 2–7 have been reported in any animal or human in China, concern has been raised by the recent isolation of four new viruses (Aravan [10], Khujand [11], Irkut [12], and West Caucasian bat viruses [13]) from bats in Eurasia. These have been incorporated as putative species within the lyssaviruses Genus in the Virus Taxonomy VIIIth Report [14], and will thus probably eventually become lyssavirus genotypes 8–11. More recent, new lyssaviruses were found in Shimoni bat [15], Natterer’s bat [16], and in African civet [17]. Probable lyssavirus infections have also been discovered in the sera of bat specimens from several countries in Southeast Asia, namely the Philippines, Cambodia, Thailand, and Bangladesh [18–21]. Virus neutralizing activity of African fruit bat (Eidolon helvum) sera against emerging lyssaviruses was confirmed [22]. These samples indicated stronger neutralizing activity against bat viruses from Eurasian and ABLV than other lyssaviruses, but it was not possible to isolate the virus, or to detect the viral genome in any of these investigations.
Yet the amount of knowledge that is currently available on bat lyssaviruses in Asia is still severely limited, mainly because of the absence of any effective surveillance system. It was in order to start to address this deficiency in our knowledge that we performed an extensive survey of bat populations in the Guangxi Province of southern China during the period 2003–2008, the results of this survey form the basis of this communication.
Materials and methods
Collection and preparation of bat brain samples
From 2003 to 2008, a total of 2,969 bat samples, some were dead and some were alive but fell onto ground of roosting caves or forest, were collected from different regions of Guangxi and provided by Guangxi Center for Animal Diseases Control and Prevention as part of the routine laboratory investigations for suspected cases, with permission from the Veterinary Administration of Guangxi (Table 1). The alive bats were euthanized in a container by inhalant halothane. All husbandry procedures were conducted in compliance with the Animal Welfare Act and the Guide for the Care and Use of Laboratory Animals. The bat brain tissues were homogenized under ice bath conditions to a 10 % emulsion with cold sterilized Hank’s solution, divided into at least five tubes, and stored at −80 °C until analysis.
Reverse transcription and polymerase chain reaction (RT-PCR)
The total RNA of each bat brain sample (10 % emulsion) was extracted using Trizol (Nippon Gene, Japan), then the cDNAs of the N gene were synthesized by a combination of reverse transcription and polymerase chain reaction (RT-PCR), using M-MuLV reverse transcriptase (Promega) and Takara Ex Taq (Takara, Japan), respectively. Four primers previously designed by Vazquez-Moron et al. [23] were used for RT, 1-st PCR, and nested-PCR in this study. An antigenomic sense primer, 1F (5′-AAR ATN GTR GAR CAY CAC AC-3′, positions 538–557) was used for RT, and a genomic sense primer, 1R (5′-GCR TTS GAN GAR TAA GGA GA-3′, positions 892–911) was also used as a primer for 1-st PCR together with the 1F primer. A nested-PCR was performed with another pair of primers, 2F (5′-AAR ATG TGY GCI AAY TGG AG-3′, positions 574–593) and 2R (5′-TCY TGH CCI GGC TCR AAC AT-3′, positions 814–833). This latter procedure has the ability to detect 260 base pairs (bp) corresponding to nucleotides 574–833 in the N gene of the seven known lyssaviruses, including the rabies virus.
Mouse inoculation test
Adult mouse injection test
Animal experiments were performed with four-week-old Kunming mice purchased from the animal centre of Guangxi Medical University. Each mouse was cerebrally injected with 0.03 mL of supernatant of tenfold diluted samples of bat brain emulsions; four mice were used per sample. The mice were observed for 1 month post-injection and then were killed under anesthesia, and their brains were collected for the next experiments. A total of three generations of blind passages were performed and the mice’s clinical signs were observed.
Suckling mouse injection test
Litters of 1-/or 2-day-old suckling mice were purchased from the animal centre of Guangxi Medical University and were kept with their dams throughout the course of the test. The suckling mice were inoculated intracerebrally with 20 μL of supernatant of 10 % bat brain emulsions, and then observed for 2 weeks. After this time the mice brains were harvested under anesthesia and 10 % mouse brain emulsions were inoculated into new suckling mice in the same manner for up to ten passages.
Virus culture in chick embryo and in BHK-21 cells
Bat brain samples which were confirmed to be positive by RT-PCR, were inoculated into 9-day-old chick embryos from the SPF chicken farm in Beijing, and BHK-21 cells (derived from Baby hamster kidney). These were then observed for the development of lyssavirus infection.
Direct sequencing of DNA
The PCR products from bat brain samples were purified by electrophoresis on 1.2 % agarose gel, and the cDNA segment was cloned into the pMD18-T vector. Sequence analysis was carried out using the AutoCycle sequencing Kit (Pharmacia) by the ALF DNA Sequencer (Pharmacia). Sequence information from both strands was aligned and edited using the Vision X program.
Genetic analysis
Sequencing data for a total of 260 nucleotides in the N gene, corresponding to nucleotides 574–833 of ten lyssavirus isolates from bats in Guangxi were used for comparison with bat lyssaviruses available from the GenBank database. The calculation of homology of nucleotide sequences was carried out using Genetic software for Windows version 6.0.1. Alignments of homologous sequences were made with the Clustal method of the MegAlign program of the DNASTAR version 7.0 package (DNASTAR Inc., USA). A neighbor-joining (NJ) tree for all the DNA sequences was constructed using the Kimura 2-parameter model with MEGA 3.1 software [24]. Phylogenetic relationships of the lyssavirus sequences were further confirmed by maximum-likelihood in PHYML [25], and by Bayesian analysis in mrbayes-3.1.2 [26, 27], respectively. For maximum- likelihood analysis, the model of HKY85 was used [28]. The transition/transversion ratio and proportion of invariable sites were estimated by maximizing the likelihood of the phylogeny. For Bayesian analyses, four Monte Carlo Markov chains were simultaneously run for 1,200,000 generations, saving a tree every 100 generations. Posterior probability (shown as percentage) for Bayesian analyses and bootstrap values for trees were assessed to provide relative support for monophyletic groups.
Results
Survey of bat samples from Guangxi Province
Out of the total of 2,969 bats collected from 11 of the 14 regions of Guangxi, the N gene was detected in 85 of the samples (2.86 %) (Table 1) from 7 of the 11 regions: Chongzuo, Baise, Nanning, Guigang, Liuzhou, Guilin, and Hezhou.
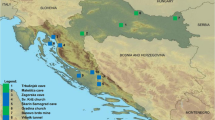
Positive samples were spread like a band from southwest to northeast Guangxi (Fig. 1), with the highest levels of N gene detection in samples from the northeast regions, Liuzhou and Guilin (6.3–7.5 %). However, there was no evidence of lyssavirus infection in samples from Hechi (northwest), Wuzhou (east), or Qinzhou, and Beihai (south).
Map of Guangxi showing locations of lyssavirus infections in bats. Infections were spread through all 6 of the identified species of bat; 17 out of 417 bats of Rhinolophidae (4.08 %), 9 of 236 bats of Hipposideridae (3.30 %), 21 of 664 bats of Vespertilionidae (3.16 %), 13 of 476 bats of Pteropodidae (2.73 %), 2 of 127 bats of Megadermatidae (1.57 %), and 4 of 287 bats of Molossidae (1.39 %). In addition, lyssavirus infection was found in 19 of 762 (2.49 %) bats whose species could not be determined
The 85 bat brain samples which were lyssavirus N gene positive by nested RT-PCR were used for mouse inoculation tests, in order to determine whether the virus could be isolated. Although no evidence of infection was found in inoculated adult mice, 21 positive samples were found with 1–2 day old suckling mice. However, none of these mice developed clinical signs during the course of the experiment, and the animals grew normally. The number of positive samples decreased progressively with each subsequent passage, but three samples were still positive after the tenth passage (Table 2).
Phylogenetic analysis based on N gene
After purifying the PCR products from the bat brain samples and cloning the cDNA segments, we compared the sequence analysis data with those for bat lyssaviruses available from the GenBank database, and calculated the homology of the nucleotide sequences. We aligned the homologous sequences and constructed a neighbor-joining (NJ) tree for all the DNA sequences, and finally confirmed the phylogenetic relationships by maximum-likelihood analyses. The transition/transversion ratio and the proportion of invariable sites were estimated by maximizing the likelihood of the phylogeny.
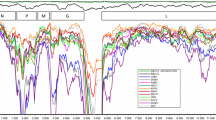
Sequencing the 260 nucleotide segment corresponding to positions 574–833 of the N gene in the ten lyssavirus isolates from Guangxi showed nucleotide homologies in the range 86.9–99.6 %. Construction of a phylogenetic tree (Fig. 2) showed that all are much more closely related to strains of the rabies virus than to other bat lyssaviruses. The strains Lipu41 and Binyang16, which could passage through suckling mice for ten generations, represent one strain with high homology with those of the Guangxi street rabies viruses GXLA, GX01; they are also closely related to the rabies viruses GXN119, GX074, and GXPL, and with SHCAN. Guigang257 and Zhongshan77 represent a second strain with high homology with ERA and PV and a close relationship to CVS-11 and HEP-flury. The other six isolates all had high homologies with one another, and were more closely related to the isolates Guigang257 and Zhongshan77 than to Lipu41 and Binyang16.
Virus detection
Thus, all ten lyssavirus isolates from bats in Guangxi bear closer resemblance to strains of the rabies virus (lyssavirus genotype 1) than to other lyssavirus genotypes. Their genes are small and could be detected by nested RT-PCR. However, all of these virus isolates replicated slowly in the suckling mice brains, and no evidence for replication was seen when these positive samples were inoculated into 4-week old mice, or chick embryo and BHK-21 cells for propagation. Incubation periods are, therefore, likely to be considerably longer than those of the classical rabies viruses, and their current risk to humans is unknown.
Discussion
The rabies virus is distributed throughout the world and has attracted attention because of its public health and economic significance. However, the other lyssaviruses have not been studied sufficiently for their potential host range and pathogenicity to be evaluated. Furthermore, there is very little information available on bat lyssaviruses in Asia, except for the identification of antibodies to ABLV in sera of Philippine bats [18], and antibodies to the rabies virus in sera of bats from southern China [9].
Since China is a country with a severe prevalence of rabies, it is an important public health issue for the background incidence levels of bat rabies or other lyssaviruses to be established. Our present study has focused on bats from the different regions of Guangxi in southern China, and results from 2,969 bats of six species collected from 2003 to 2008 show that 2.86 % were lyssavirus positive (Table 1). These positive samples were distributed in seven regions: Chongzuo, Baise, Nanning, Guigang, Liuzhou, Guilin, and Hezhou, and spread like a band from southwest to northeast Guangxi (Fig. 1). The high incidence of N gene detection in Liuzhou and Guilin (6.28 and 7.53 %, respectively) is especially worthy of note. Most investigators typically discover 1 rabid bat per 1,000 samples of dead or normal animals, however, our results showed 2.86 % positive samples. Because most of bat samples were dead and alive fell onto ground, therefore, the incidence of lyssavirus in these bat samples might be higher than that of normal bats (Table 1). Nevertheless, no antigenic positive sample was confirmed when these positive samples were further investigated by IFA using an anti-N protein monoclonal antibody with reactivity for rabies and related viruses.
When the N gene positive samples were intracerebrally injected into 1/2dayold suckling mice, 21 of 85 samples showed positive by nested RT-PCR in the first passage. The number decreased progressively with increasing number of passages, but three were still detected as positive after the tenth passage (Table 2). It is important to note that the viruses replicated slowly in the suckling mice brains and that the virus genes are small and can be detected by nested RT-PCR. Also no evidence for replication was seen when these positive samples were inoculated into chick embryo and BHK-21 cells for propagation.
According to the nucleotide sequence analyses based on a 260 nucleotide segment of positions 574–833 of the N gene, the two isolates Lipu41 and Binyang16 differed from the other eight isolates, and were close to the SHCAN and street rabies strains from Guangxi. The other eight isolates were closer to the fixed strains ERA, PV, CVS-11, and HEP-flury. All 10 positive samples from Guangxi were distinctly different from other bat viruses, such as EBLV-1 and EBLV-2 from west Europe; Aravan virus, Khujand virus, ABLV, Irkut virus, West Caucasian bat virus from east Europe; LBV, MOKV and Duvenhuge virus from Africa; and bat ABLV from Australian (Table 3).
These results provide an indication of the diversity of the strains of the rabies virus that exists even in close communities, such as bats in Guangxi. Consistent with previous studies on the evolution of lyssaviruses [29], the strains of the rabies viruses found in Guangxi bats also show a distinct geographical distribution according to the sequencing analysis. Thus continued vigilance of wild animal populations is necessary because of the rapid rate at which such viral mutations can occur, and the possibility of the production of strains with the ability to cross species boundaries [30].
This research provides an initial insight into the biological properties of the rabies virus in bat populations of southern China, and represents a foundation on which to build an informed assessment of the potential risk of bats as vectors for transmission of infectious diseases to humans, which also include the emerging viral zoonoses, due to Ebola or Marburg virus [31], Nipah virus [32], Coronaviruses [33], and Avian influenza virus [34]. Rabies encephalitis is known as a potential risk for transmission by bats [35], but except for one case of rabies following a bat bite [36], Asia is at present the only continent where bat-transmitted human rabies is not currently officially recognized as a problem. However, this may be in part the consequence of a lack of appropriate reporting. It is now widely believed that bat rabies is present throughout the world, and may have been present for a long time [37]. Therefore, since bats represent a reservoir for the maintenance and evolution of rabies and other lyssavirus genotypes, it is important to devote increased attention to evaluating their current status and predicting the potential future role of these mammals in disease development. In particular, increased efforts are needed to explore the differences of replication, genetic, and pathogenic features between the classical rabies virus and other lyssaviruses, especially since there has been a recent report of rapid mutation of the rabies virus in wild animals in the USA [30].
It is worthy of note that the strains Lipu41 and Binyang16, which could passage in suckling mice for ten generations, possessed high homologies with those of street rabies viruses GXLA, GX01, GXN119, GX074, and GXPL from Guangxi (Fig. 2). The identification of these phenomena will be helpful for providing an insight into the biological properties of rabies and other lyssaviruses.
References
H. Si, Z.M. Guo, Y.T. Hao, Y.G. Liu, D.M. Zhang, S.Q. Rao, J.H. Lu, BMC Infect. Dis. 8, 113 (2008)
L.R. Boulger, J.S. Porterfield, Trans. R. Soc. Trop. Med. Hyg. 52, 421–424 (1958)
J.B. Familusi, D.L. Moore, Afr. J. Med. Sci. 3, 93–96 (1972)
G.H. Tignor, F.A. Murphy, H.F. Clark, R.E. Shope P., Madore S.P., Bauer S.M., Buckley S.M., Meredith A.C.D., J. General Virol. 37, 565–611 (1977)
B. Amengual, J.E. Whitby, A. King, J.S. Cobo, H. Bourhy, J. General Virol. 78(Pt 9), 2319–2328 (1997)
N. Johnson, P.R. Wakeley, S.M. Brookes, A.R. Fooks, Emerg. Infect. Dis. 12, 1142–1144 (2006)
G.C. Fraser, P.T. Hooper, R.A. Lunt, A.R. Gould, L.J. Gleeson, A.D. Hyatt, G.M. Russell, J.A. Kattenbelt, Emerg. Infect. Dis. 2, 327–331 (1996)
World Health Organization technical report series 931, 1–88, back cover (2005)
Y. Jiang, L. Wang, Z. Lu, H. Xuan, X. Han, X. Xia, F. Zhao, C. Tu, Vector Borne Zoonotic Dis. 10, 177–181 (2010)
Y.T. Arai, I.V. Kuzmin, Y. Kameoka, A.D. Botvinkin, Emerg. Infect. Dis. 9, 333–337 (2003)
I.V. Kuzmin, L.A. Orciari, Y.T. Arai, J.S. Smith, C.A. Hanlon, Y. Kameoka, C.E. Rupprecht, Virus Res. 97, 65–79 (2003)
A.D. Botvinkin, E.M. Poleschuk, I.V. Kuzmin, T.I. Borisova, S.V. Gazaryan, P. Yager, C.E. Rupprecht, Emerg. Infect. Dis. 9, 1623–1625 (2003)
I.V. Kuzmin, G.J. Hughes, A.D. Botvinkin, L.A. Orciari, C.E. Rupprecht, Virus Res. 111, 28–43 (2005)
C.M. Fauquet, M.A. Mayo, J. Maniloff, U. Desselberger, L.A. Ball, Virus Taxonomy: Classification and Nomenclature of Viruses (Elsevier Academic Press, London, 2005)
I.V. Kuzmin, A.E. Mayer, M. Niezgoda, W. Markotter, B. Agwanda, R.F. Breiman, C.E. Rupprecht, Virus Res. 149, 197–210 (2010)
C.M. Freuling, M. Beer, F.J. Conraths, S. Finke, B. Hoffmann, B. Keller, J. Kliemt, T.C. Mettenleiter, E. Muhlbach, J.P. Teifke, P. Wohlsein, T. Muller, Emerg. Infect. Dis. 17, 1519–1522 (2011)
D.A. Marston, D.L. Horton, C. Ngeleja, K. Hampson, L.M. McElhinney, A.C. Banyard, D. Haydon, S. Cleaveland, C.E. Rupprecht, M. Bigambo, A.R. Fooks, T. Lembo, Emerg. Infect. Dis. 18, 664–667 (2012)
P.M. Arguin, K. Murray-Lillibridge, M.E. Miranda, J.S. Smith, A.B. Calaor, C.E. Rupprecht, Emerg. Infect. Dis. 8, 258–262 (2002)
I.V. Kuzmin, M. Niezgoda, D.S. Carroll, N. Keeler, M.J. Hossain, R.F. Breiman, T.G. Ksiazek, C.E. Rupprecht, Emerg. Infect. Dis. 12, 486–488 (2006)
B. Lumlertdacha, K. Boongird, S. Wanghongsa, S. Wacharapluesadee, L. Chanhome, P. Khawplod, T. Hemachudha, I. Kuzmin, C.E. Rupprecht, Emerg. Infect. Dis. 11, 232–236 (2005)
J.M. Reynes, S. Molia, L. Audry, S. Hout, S. Ngin, J. Walston, H. Bourhy, Emerg. Infect. Dis. 10, 2231–2234 (2004)
E. Wright, D.T. Hayman, A. Vaughan, N.J. Temperton, J.L. Wood, A.A. Cunningham, R. Suu-Ire, R.A. Weiss, A.R. Fooks, Virology 408, 183–189 (2010)
S. Vazquez-Moron, A. Avellon, J.E. Echevarria, J. Virol. Methods 135, 281–287 (2006)
S. Kumar, K. Tamura, M. Nei, Briefings Bioinform. 5, 150–163 (2004)
S. Guindon, O. Gascuel, Syst. Biol. 52, 696–704 (2003)
J.P. Huelsenbeck, F. Ronquist, R. Nielsen, J.P. Bollback, Science 294, 2310–2314 (2001)
F. Ronquist, J.P. Huelsenbeck, Bioinformatics 19, 1572–1574 (2003)
M. Hasegawa, H. Kishino, T. Yano, J. Mol. Evol. 22, 160–174 (1985)
O. Delmas, E.C. Holmes, C. Talbi, F. Larrous, L. Dacheux, C. Bouchier, H. Bourhy, PLoS ONE 3, e2057 (2008)
A. Minard, National Geographic News (2009)
Y. Suzuki, T. Gojobori, Mol. Biol. Evol. 14, 800–806 (1997)
S. Wacharapluesadee, B. Lumlertdacha, K. Boongird, S. Wanghongsa, L. Chanhome, P. Rollin, P. Stockton, C.E. Rupprecht, T.G. Ksiazek, T. Hemachudha, Emerg. Infect. Dis. 11, 1949–1951 (2005)
P.A. Rota, M.S. Oberste, S.S. Monroe, W.A. Nix, R. Campagnoli, J.P. Icenogle, S. Penaranda, B. Bankamp, K. Maher, M.H. Chen, S. Tong, A. Tamin, L. Lowe, M. Frace, J.L. DeRisi, Q. Chen, D. Wang, D.D. Erdman, T.C. Peret, C. Burns, T.G. Ksiazek, P.E. Rollin, A. Sanchez, S. Liffick, B. Holloway, J. Limor, K. McCaustland, M. Olsen-Rasmussen, R. Fouchier, S. Gunther, A.D. Osterhaus, C. Drosten, M.A. Pallansch, L.J. Anderson, W.J. Bellini, Science 300, 1394–1399 (2003)
T. Tiensin, P. Chaitaweesub, T. Songserm, A. Chaisingh, W. Hoonsuwan, C. Buranathai, T. Parakamawongsa, S. Premashthira, A. Amonsin, M. Gilbert, M. Nielen, A. Stegeman, Emerg. Infect. Dis. 11, 1664–1672 (2005)
M.C. Schneider, P.C. Romijn, W. Uieda, H. Tamayo, D.F. da Silva, A. Belotto, J.B. da Silva, L.F. Leanes, Revista panamericana de salud publica (Pan Am. J. Public Health) 25, 260–269 (2009)
N. Veeraraghavan, Item D Pasteur Institute of south India, Cocoor, annual report of the director 1953 and scientific report (1954)
A.A. King, C.D. Meredith, G.R. Thomson, Indian J. Med. Res. 187, 267–295 (1994)
Acknowledgments
We are thankful to the senior scientist Dr. Bernard A. Goodman for his kind advice and editing the manuscript. The study was supported by the Key Technology Research & Development Program (Guikegong0537008-3C) and the Natural Science Research Program of Guangxi (Guikezi2049007); and the National Basic Research Program (973-Program 2005CB523003), China.
Author information
Authors and Affiliations
Corresponding author
Additional information
Wen Wang, Wei-Li Yin, Hai-Bo Tang and Ting Rong Luo contributed equally to this study.
Rights and permissions
About this article
Cite this article
Lu, ZL., Wang, W., Yin, WL. et al. Lyssavirus surveillance in bats of southern China’s Guangxi Province. Virus Genes 46, 293–301 (2013). https://doi.org/10.1007/s11262-012-0854-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-012-0854-2