Abstract
The aim of this study was to develop a one-step real-time reverse transcription-polymerase chain reaction assay using the minor groove binding probe (MGB rRT-PCR) for rapid and quantitative detection of classical swine fever virus (CSFV). The method, which targets the 5′-nontranslated region (5′NTR) of the viral genome, detected all CSFV isolate tested, but not heterologous pathogens. Using an in vitro transcript of the 5′NTR as a quantitative standard for the CSFV genome copy number, the assay had a detection limit of 10 copies/reaction, and the standard curve had a linear range from 10 to 107 copies/reaction, with good reproducibility. As determined by an end-point dilution comparison, in most case, the sensitivity of the MGB rRT-PCR was approximately 10-fold higher than that of virus isolation and the rRT-PCR using the standard Taqman probe (standard rRT-PCR). The agreement between the MGB rRT-PCR and standard rRT-PCR, or virus isolation was 93.3% and 76.7%, respectively, when detecting 261 field samples. Due to its rapidity, high specificity and sensitivity, the MGB rRT-PCR assay provides a valuable tool for diagnosis and molecular studies of CSFV biology.
Similar content being viewed by others
Introduction
Classical swine fever virus (CSFV) is the causative agent of a highly contagious disease of demostic pigs and wild boar. CSFV belongs, together with bovine viral diarrhea virus (BVDV) and border disease virus (BDV), to the genus Pestivirus in the family Flaviviridae. CSFV is an enveloped virus with a 12.5 kb single-stranded RNA genome of positive sense (Horzinek 1991). The genome encodes a single open reading frame, which is translated in a cap-independent manner giving a single polyprotein that is co- and post-translationally processed to provide structural and non-structural proteins of the virus (Meyers et al. 1996). Both ends of the CSFV genome contain nontranslated regions (NTR), which are highly conserved among all virus isolates (Boye et al. 1991; Lowings et al. 1996; Schroeder and Balassu-Chan 1990; Wirz et al. 1993).
Classical swine fever (CSF) is one of the most devastating diseases of pigs and it poses a considerable threat to the porcine industry worldwide (Horst et al. 1999). Significant portions of the losses occur in developed countries where slaughter of infected pigs, is the preferred method for CSFV control and eradication (Moennig 2000). In some countries, the subclinical and asymptomatic infection of CSFV is universal existence, not easily recognized by farmers and veterinarians (Moennig et al. 2003; Tu 2003). Therefore, the diagnosis of CSF in a herd of pigs based on clinical signs can often be problematic. Consequently, the laboratory confirmation of suspected cases is adivisable. Although the traditional virus isolation still remains the “gold standard”, the time required for obtaining the result presents one of the major drawbacks for its use in early detection. Rapid and precise detection of CSFV is critical for disease containment.
During the last decade, a number of PCR assays were described, and proved to be a rapid, sensitive and specific diagnostic tool (Barlic-Maganja and Grom 2001; Hyndman et al. 1998; Sandvik et al. 1997; Vilcek et al. 1994). Detection of amplified PCR products by gel-based systems bears the risk of cross-contaminations, does not allow an exact quantification of genome copies (Belak and Thoren 2001). The real-time PCR assay is developed by the introduction of fluorogenic probes into the PCR system. Using this assay, quantitative detection of sequence specific templates was achieved in real-time, specificity is ensured by the hybridization reaction, and cross-contaminations are largely avoided (Gibson et al. 1996; Heid et al. 1996).
A number of detection systems are available for real-time PCR. Among them, the detection assays based on the non-specific intercalating dyes (SYBR Green I dye) and the specific hydrolyzing fluorogenic-probe (Taqman) are commonly used. A Taqman probe is a linear oligonucleotide with a fluorogenic dye attached to the 5′ end and a quencher molecule to the 3′ end. After hybridization to its complementary DNA template, it is degraded by the 5′-3′ exonuclease activity of the Taq polymerase during extension, releasing the fluorescence of the reporter dye. Recently the novel minor groove binding (MGB) probes were developed. The MGB moiety of the probe stabilizes the hybridization of the probe with single-stranded DNA targets, give the higher melting temperature, thereby allow the design of significantly shorter probes (13–18 nucleotides) that are more specific, especially if there is a mismatch in the MGB region of the duplex (Afonina et al. 2002; Kumar et al. 1998; Kutyavin et al. 2000).
For the detection of CSFV, real-time RT-PCR assays based on the standard Taqman probes (Standard rRT-PCR) have already been described (Liu et al. 2007; McGoldrick et al. 1998; Risatti et al. 2003, 2005; Wen et al. 2004; Zhao et al. 2008). In this study, a novel one-step real-time RT-PCR assay using the MGB probe (MGB rRT-PCR) was developed. The sensitivity, specificity and reproducibility of this quantitative detection method for CSFV are assessed and compared with standard rRT-PCR and virus isolation (VI) techniques.
Materials and methods
Viruses
CSFV HB0809 strain, WH1002A strain, WH1002B strain, WX0911 strain, 39 strain, porcine reproductive and respiratory syndrome virus (PRRSV) HBKM strain, Ch-1a stain, pseudorabies virus (PRV) Hubei strain, porcine circovirus type II (PCVII) HBZX stain, Japanese encephalitis virus (JEV) HB94 stain, porcine rotavirus (RV) XZ9212 strain, and porcine parvovirus (PPV) JS9303 were maintained in our laboratory. CSFV HCLV strain, Thiverval strain, Shimen strain, and BVDV AV96 strain, NADL stain are purchased from China Veterinary Culture collection Center. CSFV Henhde.98 strain, CQ2.98 strain, Gdnh1.98 strain, and Heblf3.98 strain were kindly provided by Prof. Yi-Bao Ning (National Classical Swine Fever Virus Refference Laboratory, China).
Primers and MGB probe
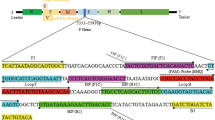
The highly conserved 5′-nontranslated region (5′NTR) of the viral genome was selected as the amplification target. Mutiple sequences were aligned using CLUSTAL W software program (Version 1.83; EBI, Cambridge). According to sequences comparison among the CSFV strains, the primers were designed as follow: CSFV-F 5′-CAG CGA CGG CCG AAC TG-3′ (location, 75-91); CSFV-R 5′-CAG GAC TTA GAC CAC CCA GGG-3′ (location, 150-170), the MGB probe: CSFV-P 5′-TCG CCA CTA CGG CTA G-3′ (location, 131-146). The MGB probe tagged with 6-carboxyfluorescein (FAM) as the 5′-reporter dye were obtained from Applied Biosystems, USA. It was expected to amplify a fragment of 96 bp.
A pair of primers for the preparation of viral RNA strandard were designed by the Primer Premier 5.0 (Applied Biosystems) as follow: 5′NTR-F 5′-TAA TAC GAC TCA CTA TA GGG CCT CCC TCC AGC GAC-3′ (location, 64-81) and 5′NTR-R 5′-CTA TCA GGT CGT ACT CCC ATC AC-3′ (location, 295-317), and were expected to amplify partial 5′NTR fragment of 254 bp (underlined nucleotides representing the T7 RNA polymerase promoter).
RNA extraction
Total RNA was extracted directly from 100 μl supernatants of infected cell cultures, whole boold or tissue homogenates, using the Trizol method according to the manufacture’s instructions (Invitrogen, GB). RNA was eluted using 50 μl of DEPC-treated water.
RNA strandard
In order to determine the absolute numer of CSFV genome copies, RNA transcripts were synthesized in vitro, and serial dilutions were used in real-time RT-PCR assays to generate external standard curves. A 254 bp PCR product amplified by the primer pair (5′NTR-F; 5′NTR-R) from the plasmid pT7SM (Accession No. AF092448), was utilized as the template for transcription. RNA standard was transcripted by the RiboMax Large Scale Production System-T7 Kit (Promega, WI, USA), using 0.2 μg PCR product as template. RNA product was treated with 2 U of DNase I (Promega, WI, USA) for 25 min at 37°C, and then were extracted with phenol-chloroform and precipitated with ethanol. The purified RNA was quantified using spectrophotometer (OD260), then stored at −70°C until use.
MGB rRT-PCR assay
MGB rRT-PCR was performed in MJ Opticon2 Real-time PCR Detection System. The concentrations of primers, probe and Mg2+ were optimized to obtain specific fluorescent signals. The assay was performed in a final volume of 20 μl containing 5 μl of RNA, 2 μl of 10× Hotstrat Taq Buffer, 3 μl of MgCl2 (25 mM), 0.4 μl of dNTPs (10 mM each), 1.5 μl of each PCR primer (10 μM), 0.4 μl of MGB probe (10 μM), 2 U of HotStart Taq polymerase (Transgen, China), and 40 U of TranScript Reverse Transcriptase (Transgen, China). Prior to amplification, the RNA was transcripted at 50°C for 30 min. This was followed by 94°C for 10 min for activation of the HotStart Taq polymerase and inactivation of reverse transcriptase. Cycle times were: 45 cycles of denaturation at 94°C for 5 sec, of annealing and extension at 60°C for 25 sec. Fluorescent signal measurements were carried out after the extension step.
Standard rRT-PCR assay
The standard rRT-PCR assay was performed as we previously described (Wen et al. 2004). In brief, the highly conserved 5′NTR was selected as amplification target. The primers and probe used in the standard rRT-PCR assay were designed and analyzed by Primer Premier 5.0. The primers used in this assay: F1 5′-CCA TGC CCA TAG TAG GAC TAG CAA A-3′ (location, 98-122); R1 5′-TCA CGT CGA ACT ACT GAC GAC TGT-3′ (location, 179-202). The Taqman probe: P1 5′-TAG CCG TAG TGG CGA GCT CCC TGG GTG GT-3′ (location, 132-160). It was labeled with FAM at the 5′ end, and TAMRA at the 3′ end.
Virus isolation assay
For isolation of CSFV, samples were inoculated in PK-15 cells at 37°C, passed twice more at 4-day intervals by transferring freeze-thawed cultures onto PK-15 cells, and followed by immunofluorescent antibody test with a CSFV-specific polyclonal anti-NS3 antibody.
Clinical samples for CSFV detection
For sensitivity test, a total of 35 clinincal samples were obtained from 5 CSFV infected pigs during the CSFV incident in Hubei province in 2007. For agreement test, 261 whole blood samples were collected from infected (54/261) or non-infected pigs (207/261) in different farms in Hubei province. Infection status was diagnosed based on clinical signs and pathological finding.
Results
Standard curve of the MGB rRT-PCR assay
The quantitative linear range of the MGB rRT-PCR assay for CSFV genomic RNA was determined by using 10-fold serial dilutions of the RNA standard (Fig. 1a and b). The values of Cycle threshold (Ct) were measured and plotted against the known copy numbers of the RNA standard dilutions. The standard curve covered a linear range of seven orders of magnitude (101 to 107 copies/reaction). Dilution curve was obtained from total RNA of CSFV-infected PK-15 cells and its amplification efficiency was similar to that of the standard curve (data not shown). The MGB rRT-PCR assay enable detection of as few as 10 CSFV genomic copies.
a Opticon2 PCR amplification and b standard curve graph illustrating 10-fold serial dilutions used to determine the sensitivity of the MGB rRT-PCR for CSFV genomic copies. Template: standard CSFV 5′NTR-RNA (in vitro transcripts); concentrations: 101 to 107 copies/reaction (from right to left). The threshold (T) of normalized reporter fluorescence used for Ct calculation is represented with a black horizontal line. Values of standard curve are the averages of three independent experiments (Mean ± SEM, n = 3)
Specificity of the MGB rRT-PCR assay
A collection of 12 archived CSFV isolates of different origin, and 9 heterologous porcine pathogens (Table 1) was tested to evaluate the specificity of the assay. As shown in Table 1, all CSFV strains tested were detected by the MGB rRT-PCR assay, while no positive signal was obtained when heterologous pathogens, including the closely related BVDV, were tested. Thus, the results indicated a high specificity for the MGB rRT-PCR assay.
Reproducibility of the MGB rRT-PCR assay
The reproducibility of MGB rRT-PCR assay was tested by detecting RNA extracted from 100-fold serial dilutions of CSFV Shimen strain and HCLV strain, and consecutively performed the assay five times. Table 2 shows the mean Ct values along with values for the coefficient of variation (CV) for each dilution in the inter-assay. The results demonstrate that the assay was highly reproducible with the CV ranging between 0.87% and 1.76% for Shimen strain and between 1.55% and 2.32% for HCLV strain.
Performance comparison among the MGB rRT-PCR, standard rRT-PCR and virus isolation assays
In order to assess whether the MGB rRT-PCR had greater sensitivity than virus isolation and Standard rRT-PCR assay as we described previously (Wen et al. 2004), different clinical samples were obtained from 5 CSFV infected pigs, and the four dilutions of each sample were tested by these assays (Table 3). The dilutions chosen cover the detection limit of these assays. One aliquot of each dilution was used for virus isolation, a second aliquot was used for RNA isolation and real-time RT-PCR analysis, results were summarized in Table 3. As shown in Table 3, the MGB rRT-PCR could detect the spleen sample at the highest dilution of 10−7, and the muscle sample at the lowest dilution of 10−2. In most case, the sensitivity of the MGB rRT-PCR was higher (spleen, kidney, tonsil, muscle, whole blood) than that of the standard rRT-PCR and virus isolation. However, in the case of serum and nasal swab sample, three assays had the comparable detection limit.
The agreements between the MGB rRT-PCR, standard rRT-PCR and virus isolation
To evaluate the diagnostic efficacy of the MGB rRT-PCR, 261 whole blood samples were tested in parallel with the virus isolation and the standard rRT-PCR assays. Out of 261 clinical samples, 54 samples were from CSFV suspected cases in field. As shown by Table 4, of 261 samples, 30 were positive by the MGB rRT-PCR assay, 28 were standard rRT-PCR positive and 23 were virus isolation positive. All the MGB rRT-PCR positive samples were further identified by PCR sequencing (data not shown). As compared to the MGB rRT-PCR, the virus isolation was found to be 76.7% (23/30) agreement, and the standard rRT-PCR was 93.3% (28/30).
Discussion
The detection of viruses by rapid and reliable techniques is still one of the most demanding tasks in veterinary diagnosis. To address the need, the real-time fluorescent quantitative PCR has been developed as a quick, sensitive, specific and high throughput methodology for quantitative detection of genetic sequences within samples. Real-time RT-PCR assays have been extensively used for the detection of ecomonically important animal virus including foot-and-mouth disease virus (Callahan et al. 2002; Reid et al. 2002), PRRSV (Kleiboeker et al. 2005; Martinez et al. 2008), swine vesicular disease virus (Reid et al. 2004), and BVDV (Bhudevi and Weinstock 2003; Letellier and Kerkhofs 2003).
Several real-time RT-PCR assays for CSFV have also been reported (Jamnikar Ciglenecki et al. 2008; McGoldrick et al. 1998; Risatti et al. 2003; Zhao et al. 2008). Evaluated with nasal swab and blood of experimentally or in contact infected swine, Risatti et al demonstrated that the assay was the same or more sensitive than virus isolation (Risatti et al. 2003). Using the clincal samples obtained from sympomatic and asymptomatic swines in the Dominican Republic, the sensitivity of this assay exceeded the diagnostic sensitivity of virus isolation with little loss of specificity (Risatti et al. 2005). Zhao et al developed a multiplex real-time RT-PCR for the quantitative and differential detection of wild-type viruses and C-strain vaccine widely used in China (Zhao et al. 2008). By using this assay, pigs infected with wild-type CSFV can be identified and removed from vaccinated population, which will help to establish a CSF-free swine herd. However, these assays were all optimized using the conventional longer Taqman probes.
Our study has validated the one-step real-time RT-PCR assay for CSFV using the novel MGB probe. Primers and a MGB probe were designed specific for a region of CSFV 5′NTR that is strictly conserved among most CSFV sequence available (Meyers and Thiel 1996). In this way, the MGB rRT-PCR assay was expected to detect CSFV belonging to all subtypes, and lineages within subtypes. The collected results further confirmed that the MGB rRT-PCR assay is highly specific for detection of all tested CSFV strains.
The real-time MGB rRT-PCR assay can be used also for a quantitative analysis. The test described here is extremely sensitive, being able to detect about 10 gene copies/reaction of in vitro transcribed RNA. Standard curves were constructed showing a dynamic range of seven orders of magnitude for transcribed RNA. Results from the sensitivity comparison test showed that, in most case, the sensitivity of the MGB rRT-PCR was approximately 10-fold higher than that of the standard rRT-PCR and virus isolation assays. This is extremely important in routine diagnostic studies, particularly when the amount of CSFV RNA in field specimens is low. The reason for the increased sensitivity of the MGB rRT-PCR assay was possibly due to the MGB moiety increases the Tm of the hybridized probe, and facilitates more efficiently binding to the target sequence. Another possible reason is the probe containing a non-fluorescent quencher that does not emit any detectable fluorescence, thus results in the lower background and greater accuracy in the measurement of reporter-specific signals.
We can also conclude from Table 4 that the virus titres in spleen, kidney, tonsil, whole blood are much higher than nasal swabs and serum, but the tissue homogenates must be treated by a complicate procedure (homogenized, frozen and thaw three times, centrifuged). Therefore, the whole blood are more suitable clinical materials for pigs to be diagnosed.
The results obtained in the examination of 261 whole blood samples, collected from infected or non-infected farms in Hubei provine of China, demonstrated that the developed MGB rRT-PCR assay can be useful for the rapid and sensitive diagnosis of CSFV. The diagnostic sensitivity of the MGB rRT-PCR assay (100%, 30/30) was higher than that of standard rRT-PCR (93.3%, 28/30), and virus isolation assay (76.7%, 23/30). ALL MGB rRT-PCR positive samples were further confirmed by PCR sequencing. Compared to virus isolation assay, the important characterization of MGB rRT-PCR is its rapidity in producing results. The assay developed in this study is performed in less than 2.5 h, while virus isolation is performed in serveral days. This is a large advantage when speed is desired, for instance in an outbreak of disease in a herd of valuable livestock.
In summary, this work described a rapid, specific, highly sensitive and throughput MGB rRT-PCR assay for detection of CSFV in clinical samples. Although further validation of the techniques using a wide variety of clinical samples, containing different subtype of virus, would be essential, the results obtained in this study using clinical material show the MGB rRT-PCR assay as useful tool for molecular diagnosis of CSFV.
References
Afonina IA, Reed MW, Lusby E, Shishkina IG, and Belousov YS (2002) Minor groove binder-conjugated DNA probes for quantitative DNA detection by hybridization-triggered fluorescence. Biotechniques 32(4): 940–944
Barlic-Maganja D, and Grom J (2001) Highly sensitive one-tube RT-PCR and microplate hybridisation assay for the detection and for the discrimination of classical swine fever virus from other pestiviruses. J Virol Methods 95(1–2): 101–110
Belak S, and Thoren P (2001) Molecular diagnosis of animal diseases: some experiences over the past decade. Expert Rev Mol Diagn 1(4): 434–443
Bhudevi B, and Weinstock D (2003) Detection of bovine viral diarrhea virus in formalin fixed paraffin embedded tissue sections by real time RT-PCR (Taqman). J Virol Methods 109(1): 25–30
Boye M, Kamstrup S, and Dalsgaard K (1991) Specific sequence amplification of bovine virus diarrhea virus (BVDV) and hog cholera virus and sequencing of BVDV nucleic acid. Vet Microbiol 29(1): 1–13
Callahan JD, Brown F, Osorio FA, Sur JH, Kramer E, Long GW, Lubroth J, Ellis SJ, Shoulars KS, Gaffney KL, Rock DL, and Nelson WM (2002) Use of a portable real-time reverse transcriptase-polymerase chain reaction assay for rapid detection of foot-and-mouth disease virus. J Am Vet Med Assoc 220(11): 1636–1642
Gibson UE, Heid CA, and Williams PM (1996) A novel method for real time quantitative RT-PCR. Genome Res 6(10): 995–1001
Heid CA, Stevens J, Livak KJ, and Williams PM (1996) Real time quantitative PCR. Genome Res 6(10): 986–994
Horst HS, Meuwissen MP, Smak JA, and Van der Meijs CC (1999) The involvement of the agriculture industry and government in animal disease emergencies and the funding of compensation in western Europe. Rev Sci Tech 18(1): 30–37
Horzinek MC (1991) Pestiviruses--taxonomic perspectives. Arch Virol Suppl 3: 1–5
Hyndman L, Vilcek S, Conner J, and Nettleton PF (1998) A novel nested reverse transcription PCR detects bovine viral diarrhoea virus in fluids from aborted bovine fetuses. J Virol Methods 71(1): 69–76
Jamnikar Ciglenecki U, Grom J, Toplak I, Jemersic L, and Barlic-Maganja D (2008) Real-time RT-PCR assay for rapid and specific detection of classical swine fever virus: comparison of SYBR Green and TaqMan MGB detection methods using novel MGB probes. J Virol Methods 147(2): 257–264
Kleiboeker SB, Schommer SK, Lee SM, Watkins S, Chittick W, and Polson D (2005) Simultaneous detection of North American and European porcine reproductive and respiratory syndrome virus using real-time quantitative reverse transcriptase-PCR. J Vet Diagn Invest 17(2): 165–170
Kumar S, Reed MW, Gamper HB, Jr Gorn VV, Lukhtanov EA, Foti M, West J, Meyer RB, Jr and Schweitzer BI (1998) Solution structure of a highly stable DNA duplex conjugated to a minor groove binder. Nucleic Acids Res 26(3):831–838
Kutyavin IV, Afonina IA, Mills A, Gorn VV, Lukhtanov EA, Belousov ES, Singer MJ, Walburger DK, Lokhov SG, Gall AA, Dempcy R, Reed MW, Meyer RB, and Hedgpeth J (2000) 3′-minor groove binder-DNA probes increase sequence specificity at PCR extension temperatures. Nucleic Acids Res 28(2): 655–661
Letellier C, and Kerkhofs P (2003) Real-time PCR for simultaneous detection and genotyping of bovine viral diarrhea virus. J Virol Methods 114(1): 21–27
Liu L, Widen F, Baule C, and Belak S (2007) A one-step, gel-based RT-PCR assay with comparable performance to real-time RT-PCR for detection of classical swine fever virus. J Virol Methods 139(2): 203–207
Lowings P, Ibata G, Needham J, and Paton D (1996) Classical swine fever virus diversity and evolution. J Gen Virol 77 ( Pt 6): 1311–1321
Martinez E, Riera P, Sitja M, Fang Y, Oliveira S, and Maldonado J (2008) Simultaneous detection and genotyping of porcine reproductive and respiratory syndrome virus (PRRSV) by real-time RT-PCR and amplicon melting curve analysis using SYBR Green. Res Vet Sci 85(1): 184–193
McGoldrick A, Lowings JP, Ibata G, Sands JJ, Belak S, and Paton DJ (1998) A novel approach to the detection of classical swine fever virus by RT-PCR with a fluorogenic probe (TaqMan). J Virol Methods 72(2): 125–135
Meyers G, and Thiel HJ (1996). Molecular characterization of pestiviruses. Adv Virus Res 47: 53–118
Meyers G, Thiel HJ, and Rumenapf T (1996) Classical swine fever virus: recovery of infectious viruses from cDNA constructs and generation of recombinant cytopathogenic defective interfering particles. J Virol 70(3): 1588–1595
Moennig V (2000) Introduction to classical swine fever: virus, disease and control policy. Vet Microbiol 73(2-3): 93–102
Moennig V, Floegel-Niesmann G, and Greiser-Wilke I (2003) Clinical signs and epidemiology of classical swine fever: a review of new knowledge. Vet J 165(1): 11–20
Reid SM, Ferris NP, Hutchings GH, Zhang Z, Belsham GJ, and Alexandersen S (2002) Detection of all seven serotypes of foot-and-mouth disease virus by real-time, fluorogenic reverse transcription polymerase chain reaction assay. J Virol Methods 105(1): 67–80
Reid SM, Ferris NP, Hutchings GH, King DP, and Alexandersen S (2004) Evaluation of real-time reverse transcription polymerase chain reaction assays for the detection of swine vesicular disease virus. J Virol Methods 116(2): 169–176
Risatti GR, Callahan JD, Nelson WM, and Borca MV (2003) Rapid detection of classical swine fever virus by a portable real-time reverse transcriptase PCR assay. J Clin Microbiol 41(1): 500–505
Risatti G, Holinka L, Lu Z, Kutish G, Callahan JD, Nelson WM, Brea Tio E, and Borca MV (2005) Diagnostic evaluation of a real-time reverse transcriptase PCR assay for detection of classical swine fever virus. J Clin Microbiol 43(1): 468–471
Sandvik T, Paton DJ, and Lowings PJ (1997) Detection and identification of ruminant and porcine pestiviruses by nested amplification of 5′ untranslated cDNA regions. J Virol Methods 64(1): 43–56
Schroeder BA, and Balassu-Chan TC (1990) Specific sequence amplification of bovine viral diarrhoea virus nucleic acid. Arch Virol 111(3–4): 239–246
Tu CC (2003) Classical Swine Fever: International Trend, Chinese Status and Control Measures. Chinese Scientia Agricultura Sinica 36(8): 955–960
Vilcek S, Herring AJ, Herring JA, Nettleton PF, Lowings JP, and Paton DJ (1994) Pestiviruses isolated from pigs, cattle and sheep can be allocated into at least three genogroups using polymerase chain reaction and restriction endonuclease analysis. Arch Virol 136(3–4): 309–323
Wen GY, Wan C, Pan ZS, and Zhang CY (2004) Rapid quantitative detection of Classical swine fever virus by real-time Taqman assay. J. Wuhan Univ. (Nat. Sci. Ed) 50: 746–750
Wirz B, Tratschin JD, Muller HK, and Mitchell DB (1993) Detection of hog cholera virus and differentiation from other pestiviruses by polymerase chain reaction. J Clin Microbiol 31(5): 1148–1154
Zhao JJ, Cheng D, Li N, Sun Y, Shi Z, Zhu QH, Tu C, Tong GZ and Qiu HJ (2008) Evaluation of a multiplex real-time RT-PCR for quantitative and differential detection of wild-type viruses and C-strain vaccine of Classical swine fever virus. Vet Microbiol 126(1–3): 1–10
Acknowledgements
The author would like to thank Dr. Zishu Pan (State Key Laboratory of Virology, Wuhan University) for kindly providing virus antibody and constructive criticisms of the manuscript. This work was supported by grant from Hubei key laboratory of Animal Embryo and Molecular Breeding (2008ZD08).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wen, G., Yang, J., Luo, Q. et al. A one-step real-time reverse transcription-polymerase chain reaction detection of classical swine fever virus using a minor groove binding probe. Vet Res Commun 34, 359–369 (2010). https://doi.org/10.1007/s11259-010-9363-8
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11259-010-9363-8