Abstract
Conditional gene knockout technology is a powerful tool to study the function of a gene in a specific tissue, organ or cell lineage. The most commonly used procedure applies the Cre-LoxP strategy, where the choice of the Cre driver promoter is critical to determine the efficiency and specificity of the system. However, a considered choice of an appropriate promoter does not always protect against the risk of unwanted recombination and the consequent deletion of the gene in other tissues than the desired one(s), due to phenomena of non-specific activation of the Cre transgene. Furthermore, the causes of these phenomena are not completely understood and this can potentially affect every strain of Cre-mice. In our study on the deletion of a same gene in two different tissues, we show that the incidence rate of non-specific recombination in unwanted tissues depends on the Cre driver strain, ranging from 100 %, rendering it useless (aP2-Cre strain), to ~5 %, which is still compatible with their use (RIP-Cre strain). The use of a simple PCR strategy conceived to detect this occurrence is indispensable when producing a tissue-specific knockout mouse. Therefore, when choosing the Cre-driver promoter, researchers not only have to be careful about its tissue-specificity and timing of activation, but should also include a systematical screening in order to exclude mice in which atypical recombination has occurred and to limit the unnecessary use of laboratory animals in uninterpretable experiments.



Similar content being viewed by others
References
Abel ED, Peroni O, Kim JK, Kim YB, Boss O, Hadro E, Minnemann T, Shulman GI, Kahn BB (2001) Adipose-selective targeting of the GLUT4 gene impairs insulin action in muscle and liver. Nature 409:729–733. doi:10.1038/35055575
Bouabe H, Okkenhaug K (2013) Gene targeting in mice: a review. Methods Mol Biol Clifton NJ 1064:315–336. doi:10.1007/978-1-62703-601-6_23
Donehower LA, Harvey M, Vogel H, McArthur MJ, Montgomery CA, Park SH, Thompson T, Ford RJ, Bradley A (1995) Effects of genetic background on tumorigenesis in p53-deficient mice. Mol Carcinog 14:16–22
Dor Y, Brown J, Martinez OI, Melton DA (2004) Adult pancreatic beta-cells are formed by self-duplication rather than stem-cell differentiation. Nature 429:41–46. doi:10.1038/nature02520
Friedel RH, Wurst W, Wefers B, Kühn R (2011) Generating conditional knockout mice. Methods Mol Biol Clifton NJ 693:205–231. doi:10.1007/978-1-60761-974-1_12
Harno E, Cottrell EC, White A (2013) Metabolic pitfalls of CNS Cre-based technology. Cell Metab 18:21–28. doi:10.1016/j.cmet.2013.05.019
Harvey M, McArthur MJ, Montgomery CA, Bradley A, Donehower LA (1993) Genetic background alters the spectrum of tumors that develop in p53-deficient mice. FASEB J Off Publ Fed Am Soc Exp Biol 7:938–943
He W, Barak Y, Hevener A, Olson P, Liao D, Le J, Nelson M, Ong E, Olefsky JM, Evans RM (2003) Adipose-specific peroxisome proliferator-activated receptor gamma knockout causes insulin resistance in fat and liver but not in muscle. Proc Natl Acad Sci USA 100:15712–15717. doi:10.1073/pnas.2536828100
Heffner CS, Herbert Pratt C, Babiuk RP, Sharma Y, Rockwood SF, Donahue LR, Eppig JT, Murray SA (2012) Supporting conditional mouse mutagenesis with a comprehensive cre characterization resource. Nat Commun 3:1218. doi:10.1038/ncomms2186
Herrera PL (2000) Adult insulin- and glucagon-producing cells differentiate from two independent cell lineages. Dev Camb Engl 127:2317–2322
Kobayashi Y, Hensch TK (2013) Germline recombination by conditional gene targeting with Parvalbumin-Cre lines. Front Neural Circuits 7:168. doi:10.3389/fncir.2013.00168
Kwan K-M (2002) Conditional alleles in mice: practical considerations for tissue-specific knockouts. Genes N Y N 2000 32:49–62
Lee KY, Russell SJ, Ussar S, Boucher J, Vernochet C, Mori MA, Smyth G, Rourk M, Cederquist C, Rosen ED, Kahn BB, Kahn CR (2013) Lessons on conditional gene targeting in mouse adipose tissue. Diabetes 62:864–874. doi:10.2337/db12-1089
Leneuve P, Zaoui R, Monget P, Le Bouc Y, Holzenberger M (2001) Genotyping of Cre-lox mice and detection of tissue-specific recombination by multiplex PCR. Biotechniques 31(1156–1160):1162
Magnuson MA, Osipovich AB (2013) Pancreas-specific Cre driver lines and considerations for their prudent use. Cell Metab 18:9–20. doi:10.1016/j.cmet.2013.06.011
Matthaei KI (2007) Genetically manipulated mice: a powerful tool with unsuspected caveats. J Physiol 582:481–488. doi:10.1113/jphysiol.2007.134908
Meng D-M, Wang L, Xu J-R, Yan S-L, Zhou L, Mi Q-S (2013) Fabp4-Cre-mediated deletion of the miRNA-processing enzyme Dicer causes mouse embryonic lethality. Acta Diabetol 50:823–824. doi:10.1007/s00592-011-0335-4
Mullican SE, Tomaru T, Gaddis CA, Peed LC, Sundaram A, Lazar MA (2013) A novel adipose-specific gene deletion model demonstrates potential pitfalls of existing methods. Mol Endocrinol Baltim Md 27:127–134. doi:10.1210/me.2012-1267
Postic C, Shiota M, Niswender KD, Jetton TL, Chen Y, Moates JM, Shelton KD, Lindner J, Cherrington AD, Magnuson MA (1999) Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic beta cell-specific gene knock-outs using Cre recombinase. J Biol Chem 274:305–315
Rempe D, Vangeison G, Hamilton J, Li Y, Jepson M, Federoff HJ (2006) Synapsin I Cre transgene expression in male mice produces germline recombination in progeny. Genes N Y N 2000 44:44–49. doi:10.1002/gene.20183
Schmidt-Supprian M, Rajewsky K (2007) Vagaries of conditional gene targeting. Nat Immunol 8:665–668. doi:10.1038/ni0707-665
Simmen FA (1994) Transgenesis in creatures great and small Transgenic Animal Technology. A laboratory handbook Edited by Carl A. Pinkert San Diego: Academic Press, Inc. (1994). 364 pp. Theriogenology 42:213–215
Song J, Xu Y, Hu X, Choi B, Tong Q (2010) Brain expression of Cre recombinase driven by pancreas-specific promoters. Genes N Y N 2000 48:628–634. doi:10.1002/dvg.20672
Tsai PT, Hull C, Chu Y, Greene-Colozzi E, Sadowski AR, Leech JM, Steinberg J, Crawley JN, Regehr WG, Sahin M (2012) Autistic-like behaviour and cerebellar dysfunction in Purkinje cell Tsc1 mutant mice. Nature 488:647–651. doi:10.1038/nature11310
Wang X (2009) Cre transgenic mouse lines. Methods Mol Biol Clifton NJ 561:265–273. doi:10.1007/978-1-60327-019-9_17
Wicksteed B, Brissova M, Yan W, Opland DM, Plank JL, Reinert RB, Dickson LM, Tamarina NA, Philipson LH, Shostak A, Bernal-Mizrachi E, Elghazi L, Roe MW, Labosky PA, Myers MG, Gannon M, Powers AC, Dempsey PJ (2010) Conditional gene targeting in mouse pancreatic ß-Cells: analysis of ectopic Cre transgene expression in the brain. Diabetes 59:3090–3098. doi:10.2337/db10-0624
Yukawa K, Kikutani H, Inomoto T, Uehira M, Bin SH, Akagi K, Yamamura K, Kishimoto T (1989) Strain dependency of B and T lymphoma development in immunoglobulin heavy chain enhancer (E mu)-myc transgenic mice. J Exp Med 170:711–726
Zeller A, Crestani F, Camenisch I, Iwasato T, Itohara S, Fritschy JM, Rudolph U (2008) Cortical glutamatergic neurons mediate the motor sedative action of diazepam. Mol Pharmacol 73:282–291. doi:10.1124/mol.107.038828
Zhang J, Dublin P, Griemsmann S, Klein A, Brehm R, Bedner P, Fleischmann BK, Steinhäuser C, Theis M (2013) Germ-line recombination activity of the widely used hGFAP-Cre and nestin-Cre transgenes. PLoS ONE 8:e82818. doi:10.1371/journal.pone.0082818
Zhu Y, Romero MI, Ghosh P, Ye Z, Charnay P, Rushing EJ, Marth JD, Parada LF (2001) Ablation of NF1 function in neurons induces abnormal development of cerebral cortex and reactive gliosis in the brain. Genes Dev 15:859–876. doi:10.1101/gad.862101
Acknowledgments
We thank Dr. Pedro Herrera for kindly providing us with RIP-Cre mice. V.S. was supported by a grant from the Fondation pour la Recherche Médicale (FRM) Grant FDT20140930804. This work was supported by Grants from Région Nord-Pas de Calais, FEDER, INSERM, A.N.R. (FXREn), Société Francophone du Diabète (SFD), Université Lille 2, Université Lille Nord de France and European Genomic Institute for Diabetes (EGID, ANR-10-LABX-46) and European Commission. B.S. is a member of the Institut Universitaire de France.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1
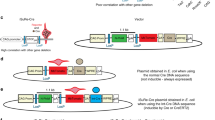
Breeding scheme used to obtain AT-POI-KO mice and their corresponding controls. POIfl/fl aP2-Cre−/− mice were crossed with POIwt/wt aP2-Cre+/? mice to generate POIfl/wt aP2-Cre−/− and POIfl/wt aP2-Cre+/− offspring. Next, POIfl/fl aP2-Cre−/− and POIfl/wt aP2-Cre+/− mice were bred together to generate POIfl/fl aP2-Cre+/− (AT-POI-KO) and POIfl/fl aP2-Cre−/− littermate mice (control mice) (TIFF 10960 kb)
Fig. S2
The null allele is present in all aP2-Cre+/− obtained from 1st cross. a, Percentages of mice from the 1st cross distributed by tail genotype for AT-POI-KO breeding. b, Representative gel showing PCR products for POI-amplified locus on genomic DNA extracted from tail biopsy of AT-POI-KO mice (TIFF 13998 kb)
Fig. S3
The null allele is present in all aP2-Cre+/− mice obtained from 2nd cross. a, Percentages of mice from the 2nd cross distributed by tail genotype from AT-POI-KO breeding. b, Representative gel showing PCR products for POI-amplified locus on genomic DNA extracted from tail biopsies of AT-POI-KO mice (TIFF 14170 kb)
Rights and permissions
About this article
Cite this article
Spinelli, V., Martin, C., Dorchies, E. et al. Screening strategy to generate cell specific recombination: a case report with the RIP-Cre mice. Transgenic Res 24, 803–812 (2015). https://doi.org/10.1007/s11248-015-9889-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11248-015-9889-1