Abstract
A C4(V3)6 multiepitopic protein was designed in an effort to pursue broad immunization against the human immunodeficiency virus (HIV). This C4(V3)6 chimeric protein is based on sequences of gp120, including epitopes from the fourth conserved domain (C4) and six tandem repeats of the third variable domain (V3), which represent different HIV isolates. The histidine-tagged C4(V3)6 was subsequently over-expressed in a recombinant Escherichia coli strain, and purified by immobilized metal ion affinity chromatography. Expression of the C4(V3)6 in both tobacco and lettuce plants was also achieved with no toxic effects on plant growth as transgenic plants were phenotypically normal. Moreover, the functional C4(V3)6 protein showed HIV antigenic determinants. The implications of these findings on the development of a new low-cost HIV vaccine are discussed.
Similar content being viewed by others
Introduction
The development of an effective vaccine against the human immunodeficiency virus (HIV) that causes acquired immune deficiency syndrome (AIDS) remains a priority in global health initiatives. However, this has been a challenge as current vaccination strategies that prevent viral infections such as those for smallpox, hepatitis B, and polio have been ineffective in preventing HIV infection. This is primarily attributed to the rapid mutation ability of the virus which facilitates viral elusiveness from the host’s cellular and humoral immune responses (Nabel et al. 2002; Smith et al. 2012).
Among several approaches explored to date against HIV, synthetic peptides have demonstrated to be promising components of HIV vaccine formulations as these are designed to include specific epitopes (Rerks-Ngarm et al. 2009; Vasan and Michael 2012). This allows for immunization against non-immunodominant epitopes, as well as those epitopes surrounded by several non-protecting antigens (Fernández et al. 1998). Previous studies have demonstrated that humoral and cellular immune responses can be elicited by both liposomes and immunostimulatory complexes (ISCOMs) formulated with HIV-1 peptides when an appropriate adjuvant is used (Agrawal et al. 2003; Staats et al. 1996; Bradney et al. 2002). Other effective approaches are based on chemical coupling of these peptides to a wide variety of carriers (Hamajima et al. 1995; Golding et al. 2002). Interestingly, polyvalent synthetic peptides have proven to be immunogenic, leading to both humoral and cellular responses in both mice and non-human primates (Hart et al. 1990, 1991). Moreover, these HIV peptide-based candidates have shown to induce neutralizing antibodies, of broad spectrum, against primary isolates (Haynes et al. 1993).
In this context, it is important to point out that, oftenly, synthetic peptides tend to be weak immunogens, and are chemically unstable. Thus, coupling a carrier to a synthetic peptide is essential for enhancing stability and increasing immunogenicity of such a peptide (Haynes et al. 2005). In addition, large-scale synthesis of long peptides is not economically feasible for large scale vaccination programs, especially in the developing world. Hence, recombinant proteins serve as viable alternatives. Through genetic engineering efforts, not only it has been possible to produce recombinant proteins, but also to design specific protein configurations containing several neutralizing epitopes in a single formulation (Lu et al. 2000; Malm et al. 2005). This allows for production of larger amino acid sequences that are likely to be immunogenic and less expensive than their synthetic counterparts. The inclusion of a number of protective epitopes within a single molecule may elicit appropriate cellular and humoral responses that can overcome evasive problems of viral mutations.
Previously, we have reported on the production and immunological characterization of a rC4V3 hybrid polypeptide carrying sequences from the fourth conserved (C4) and the third variable (V3) domains of the glycoprotein gp120MN, as this polypeptide is a strong elicitor of anti-HIV antibodies at systemic and mucosal compartments without the need for adjuvants (Varona-Santos et al. 2006). More recently, this functional polypeptide has been produced into tobacco chloroplasts, and transplastomic tobacco carrying this peptide has proven to elicit oral immunogenicity in test mice (Rubio-Infante et al. 2012). Thus, using this C4V3 polypeptide configuration to develop new molecules eliciting enhanced immune responses is worthy of consideration in further studies. In this study, we have expanded the configuration of this polypeptide to include five additional V3 loop regions from different HIV isolates to yield a protein molecule designated as C4(V3)6. This molecule was initially produced in recombinant E. coli and subsequently in transgenic both tobacco and lettuce plants in an effort to develop a low-cost production/delivery system (Rosales-Mendoza et al. 2012). The antigenic properties of the C4(V3)6 protein are also presented herein.
Materials and methods
Gene design and vector construction
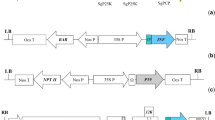
To construct the C4(V3)6 gene for E. coli-based expression, the C4V3 encoding gene reported by Varona-Santos et al. (2006) was used as backbone. An additional region encodes for five V3 loops from HIV isolates CC, RF, MN, RU, and IIIB, were designed along with linkers between each V3 epitope. This coding sequence was defined based on codon optimization performed by Genscript (USA) to aid in efficient gene translation in E. coli and cloned in frame into the C4V3 open reading frame following standard restriction and ligation reactions. Subsequently, the 900 bp full length synthetic gene was amplified by PCR. The primers used during this amplification introduced the Nco I and Hind III flanking restriction sites to facilitate further cloning steps. The PCR product was cloned into pGEM® T-Easy vector System (Promega, http://www.promega.com) and its sequence determined. Finally, the synthetic C4(V3)6 gene was cloned into the pTrcHis2C vector (Invitrogen, http://www.invitrogen.com) through the Nco I and Hind III sites to obtain the pC4(V3)6 expression vector. A positive clone was selected by restriction analysis and used for protein expression assays.
To produce an E. coli-derived protein useful for antibody analyses, a chimeric protein comprising the mouse dihydrofolate reductase (DHFR) and the V3 loop from IIIB HIV (DHFR::IIIB) was produced. PCR analysis was conducted to isolate the corresponding IIIB-V3 coding region from a full length gp120 cDNA. The amplicon was cloned in-frame at the 3’ end of the DHFR from the pQE-40 vector (Qiagen, http://www.qiagen.com) to yield the pQ40-IIIB construct.
To develop a C4(V3)6-plant expression vector, a C4(V3)6 encoding gene was synthesized (GenScript, http://www.genscript.com) following codon optimization for plant expression, and including Xba I and Sac I flanking restriction sites. The C4(V3)6 gene was cloned downstream of the CaMV 35S promoter of the pBI121 plant expression vector. Both pBI121 and the plasmid carrying the C4(V3)6 gene were digested with Xba I and Sac I endonucleases. The vector and insert of interest were ligated to construct the expression vector named pBin-C4(V3)6. Then, the ligation mix was used to transform chemically competent E. coli cells. A positive clone was selected by restriction analysis, and used for plasmid isolation. After confirming sequence integrity by standard sequencing, the construct was then mobilized into Agrobacterium tumefaciens GV3101 strain by electroporation, and this recombinant strain was used for plant transformation.
Production and purification of E. coli-derived chimeric proteins
Cells of E. coli TOP10 (Invitrogen, http://www.invitrogen.com) strain transformed with either pBin-C4(V3)6 or pQ40-IIIB expression vectors, were grown in LB medium supplemented with 100 μg/ml ampicillin. Expression was induced by adding isopropyl-thiogalactoside (IPTG) to a 1 mM final concentration. Purification of the corresponding chimeric proteins was performed by Ni-IMAC following the manufacter’s instructions (QIAGEN, http://www.qiagen.com). Briefly, bacterial cells were pelleted, resuspended in cold TE buffer (10 mM Tris–HCl, 1 mM EDTA, pH 8), and then lysed by sonication (Fisher Sonic Dismembrator Model 300). Inclusion bodies were pelleted by centrifugation at 8,000×g for 10 min, washed once with 0.5 M NaCl-1 %, Tween-20, and then twice with TE buffer. Inclusion bodies were solubilized in Urea buffer (100 mM NaH2PO4, 8 M Urea, 0.5 M NaCl, 20 mM Imidazol, pH 8) and incubated with NiNTA resin by 1 h (QIAGEN, http://www.qiagen.com). After washing the resin, recombinant proteins were eluted with binding buffer supplemented with 250 mM imidazol. In order to desalt and re-fold the recombinant proteins, elution fractions were extensively dialyzed in refolding buffer overnight (30 mM Na2CO3, 70 mM NaHCO3, 7 % sucrose, pH 9.6). The recombinant protein was clarified by centrifugation and stored at −70 °C until used. After determining the protein concentration (Bradford 1976), purity was visualized by SDS-PAGE. For C4(V3)6, endotoxin levels were assessed using an E-Toxate kit (Sigma, http://www.sigmaaldrich.com), with a limit of sensitivity of 0.05–0.1 endotoxin units (EU)/ml, following manufacturer’s instructions.
Transformation of tobacco and lettuce
The culture medium consisted of the standard Murashige and Skoog (MS) (Phytotechnology Lab, http://www.phytotechlab.com) and solidified with 0.8 % agar. Cultures were maintained at 25 °C under 16 h photoperiod (100 μmol m−2 s−1) throughout culture.
Tobacco plants carrying the C4(V3)6 gene were obtained by A. tumefaciens-mediated transformation following the standard transformation protocol of Horsch et al. (1985). Briefly, leaves from in vitro-germinated tobacco seedlings (Nicotiana tabacum cv. Petite Havana SR1) were cut into small segments, and co-cultivated with recombinant A. tumefaciens strain GV3101 carrying the pBin-C4(V3)6 vector. Following transfer of co-cultivated leaf sections to MS medium containing 200 mg/L kanamycin for selection, regenerated shoots were excised, and rooted onto a plant growth regulator (PGR)-free medium, but containing 100 mg/L of kanamycin. Successfully rooted plantlets were then transplanted into small plastic pots (15 cm in diameter) containing soil mix, placed in a large plastic tray covered with a plastic dome to maintain high humidity, and kept at a temperature of 25 ± 1 °C. One week post-transplantation, the plastic cover was gradually removed over a period of 5 days. After 2 weeks, plants were transferred to larger plastic pots (25 cm in diameter), and grown under the same environmental conditions until maturity. Seeds were collected from these plants.
Transgenic lettuce plants were obtained following using an Agrobacterium-mediated lettuce transformation reported by Curtis (2006), but with some modifications. Seeds of lettuce cv. Green Wave (Takii Seed Co. Ltd., http://www.takii.com) were germinated on MS medium. Cotyledons of 7-day-old seedlings were excised, wounded several times with a scalpel, and dipped into bacterial suspension of A. tumefaciens, derived from overnight-grown culture diluted 1:10 with LB media to a final OD600 nm of =0.2. Cotyledons were blotted dry onto a sterilized paper, and co-cultured for 2 days on a co-cultivation medium. The medium consisted of MS salts and vitamins, supplemented with 0.05 mg/L naphthaleneacetic acid (NAA), 0.25 mg/L kinetin, 2.0 mg/L 2,4-dichlorophenoxyacetic acid (2,4-D), and 2.0 g/L casein. Following co-cultivation, explants were rinsed with sterilized water, blotted dry, and placed on selection medium 1 [MS salts and vitamins, supplemented with 1.0 mg/L NAA, 0.5 mg/L 6-benzyladenine (BA), 100 mg/L kanamycin, and 500 mg/L cefotaxime]. Cotyledons were subcultured biweekly onto selection medium 1 for a period of 1 month, and then transferred to selection medium 2 [MS salts and vitamins, supplemented with 0.05 mg/L NAA, 0.2 mg/L 6-benzyladenine (BA), 100 mg/L kanamycin, and 250 mg/L cefotaxime]. Explants were subcultured biweekly. When developing kanamycin-resistant shoots reached 1 cm in length, they were transferred to rooting medium [MS salts and vitamins supplemented with 50 mg/L kanamycin and 250 mg/L cefotaxime]. Transplantation of the regenerated lettuce plants was performed as described above for tobacco.
PCR analysis
Total DNA was isolated from leaves of both putative transformants and wild-type plants according to Dellaporta et al. (1983). A 25 μL reaction mixture contained 100 ng DNA, 1.5 mM magnesium chloride, 2.5 U Taq DNA polymerase, 1 mM dNTPs, and 1 μM of primers (sense 5′TATTCGGCTATGACTGGGCA; antisense 5′GCCAACGCTATGTCCTGAT). Cycling conditions were 94 °C for 5 min (initial denaturation); 35 cycles of 95 °C for 30 s, 56 °C for 30 s, 72 °C for 60 s, and a final extension at 72 °C for 5 min. PCR products were analyzed by electrophoresis on 1 % agarose gel.
Western blot analysis
Protein extracts were obtained by resuspending 10 mg of freeze-dried leaf tissue into 50 μL of 1X reducing loading buffer. Samples were denatured by boiling for 5 min at 95 °C, debris was eliminated by centrifugation at 12,000×g for 10 min, and SDS-PAGE was performed in 4–12 % acrylamide gels under denaturing conditions. The gel was blotted on to BioTrace PVDF membrane (Pall Corporation, http://www.pall.com). After blocking in PBST plus 1 % fat-free milk, blots were incubated with either a mouse antibody directed against the Hystidine tag included at the C-terminus (1:10,000 dilution, Genscript, http://www.genscript.com), or a pool of sera from HIV positive patients. Horseradish peroxidase-conjugated secondary antibodies (1:10,000 dilution, Sigma, http://www.sigmaaldrich.com) were applied for 2 h at room temperature. Antibody binding was detected by incubation with SuperSignal West Dura solution following the manufacturer’s instructions (Thermo Scientific, http://www.thermoscientific.com). Signal detection was performed by means of an X-ray film following standard procedures. In order to estimate the amount of protein produced in lettuce, optical densitometry analysis was performed with a Biosens SC805 documentation system, using the E. coli-derived C4(V3)6 as standard.
Results
C4(V3)6 is successfully produced in E. coli
A C4(V3)6 protein was designed to obtain a multiepitopic antigen capable of eliciting antibodies against five different V3 sequences from different isolates (Table 1). The HIV C4(V3)6 encoding gene was successfully assembled and cloned into a conventional bacterial expression vector. Following transfer of this construct into an E. coli expression strain, the expected C4(V3)6 protein was successfully over-expressed by means of isopropyl β-D-1-thiogalactopyranoside (IPTG) induction. After immobilized metal affinity chromatography (IMAC) purification and refolding processes, SDS-PAGE analysis revealed an acceptable level of purity (Fig. 1). However, an additional band of ~18 kDa was also observed.
Antigenicity of the purified E. coli-derived C4(V3)6 was assessed by Western blot using a pool of sera from HIV positive patients. A protein of 23 kDa in size was detected in this assay (Fig. 2). An additional 46 kDa band was also observed, which was likely to correspond to either dimeric form of the protein or endogenous proteins associated with C4(V3)6. Interestingly, this observation was also registered when the C4V3 was produced in E. coli as expression host (Varona-Santos et al. 2006).
Production and detection of the immunoreactive C4(V3)6 protein in E. coli. Presence of the expected recombinant C4(V3)6 was determined by Western blot analysis using different primary antibodies. Purified E. coli-derived protein detected by labeling with a pool sera from HIV+ patients. Lanes: M molecular weight marker; 1 and 2, 1 μg of rC4(V3)6; and C, negative control (10 μg of a total protein sample from WT E. coli)
C4(V3)6 is successfully produced in plant cells
To explore the viability to express the C4(V3)6 protein in plant cells, the pBin-C4(V3)6 vector carrying the construct was introduced into tobacco using Agrobacterium-mediated transformation. Two months post co-cultivation, several KanR lines were rescued, transferred to soil mix, acclimatized, and grown to maturity in the greenhouse. All plants showed no visible phenotypic alterations. Most regenerated plants were PCR-positive for the of nptII transgene, thus confirming presence of the transgene. Seeds were collected from T0 plants, and these germinated on Murashige and Skoog (MS) selection medium containing kanamycin, thus indicating these plants are transformed with C4(V3)6 construct (data not shown).
Subsequently, the C4(V3)6 cassette was introduced into lettuce. Multiple KanR calli were generated over a period of 1 month post co-cultivation, and putative transformed lettuce shoots were obtained after 3 months post co-cultivation. Plants with well-developed roots were established in plant growth regulator PGR-free medium, and a total of 14 lines were transferred to soil mix, acclimatized, and grown in the greenhouse. Presence of the nptII transgene was confirmed using PCR, showing an amplicon of the expected molecular weight of 600 bp (Fig. 3). No PCR product was amplified in reactions containing DNA from untransformed plants and wild-type (control) plants.
Detection of the nptII (kanamycin resistance) gene in transformed lettuce plants. Genomic DNA samples from either transformed lines or wild-type plants were used to demonstrate the presence of the nptII gene, which is located at the T-DNA of the pBin-C4(V3)6 vector. Lanes: M 1 kb molecular weight marker; − negative control; WT wild-type plant; + positive control (pBin-C4(V3)6); and LCV1-6, lettuce candidate lines
To investigate the ability of lettuce cells to synthesize the recombinant C4(V3)6 protein, six lines were selected to detect the recombinant protein by Western-blot assays. Labelling with either anti-His monoclonal antibody (Fig. 4) or sera from HIV+ patients (data not shown) revealed presence of an immunoreactive protein of 23 kDa. This indicated appropriate production of the expected C4(V3)6 as detected earlier in E. coli (Fig. 2). Based on band intensities of plant extracts and in comparison to those of the pure rC4(V3)6 standard, it was estimated that 1 g of freeze-dried T0 lettuce leaves contained 240 μg of the C4(V3)6 protein. Following seed production, T1 lines were successfully rescued on MS selection medium containing kanamycin. These T1 plants were confirmed to carry the C4(V3)6 transgene following PCR analysis (data not shown).
Production and detection of the immunoreactive C4(V3)6 protein in lettuce cells. Presence of the expected recombinant C4(V3)6 was determined by Western blot analysis. Lettuce-derived C4(V3)6 protein was detected by labeling with an anti-His tag antibody. Lines: WT wild-type lettuce, STD 500 ng of pure C4(V3)6 as standard, and LCV1 to 6, transgenic lettuce lines
Discussion
An ideal subunit HIV vaccine would consist of a single multiepitopic recombinant protein with high immunoprotective properties, and it would be capable of inducing a broad immune response against multiple strains (or mutants) of the invading pathogen, therefore protecting against disease development (Koff 2012). Moreover, it would be highly desirable that this vaccine is produced at low-cost, thus allowing for efficacious and economic vaccination programs to be implemented worldwide (Daniell et al. 2001; Twyman et al. 2003).
In recent years, some reports have identified the importance and viability of the use of an anti-V3 as a humoral neutralizing response (Zolla-Pazner et al. 2009; Watkins et al. 2011; Totrov et al. 2010). For example, Zolla-Pazner et al. (2011) primed rabbits with a gp120 DNA vaccine and then boosted them with chimeric gp120 proteins. Their results indicated that the immune response was dependent on a neutralizing epitope and that anti-V3 antibodies could neutralize a diverse set of V3 loops. Similar findings have also been reported by Vaine et al. (2010).
In this study, we have designed a C4(V3)6 multiepitopic protein, carrying several V3 variants along with the C4 domain of HIV, and based on the previously reported recombinant immunogenic C4V3 protein (Varona-Santos et al. 2006). It is hypothesized that this new design is capable of triggering a broader humoral response, leading to neutralization activity against several HIV isolates.
When the recombinant multiepitopic C4(V3)6 protein is expressed in E. coli, it is correctly expressed. As these results were encouraging, we explored the use of plants as systems for production of this recombinant multiepitopic protein. Early on, the C4(V3)6 was introduced into a plant expression vector and transferred into tobacco, as a test system. PCR analysis of KanR plants revealed presence of T-DNA sequences in transformed tobacco plants, and all plants were phenotypically normal. This finding suggested that this complex structural protein can be introduced into plant cells without eliciting any negative effects on plant growth and development. This finding prompted us to proceed in transferring the C4(V3)6 gene construct into an edible and safe plant system, lettuce, that does not require processing for oral delivery of this candidate subunit vaccine. To date, lettuce has been used as a plant expression host for several antigens (Kanagaraj et al. 2011; Marcondes and Hansen 2008; Matsui et al. 2011; Pniewski et al. 2011; Rosales-Mendoza et al. 2012), as it is deemed as a desirable production and delivery platform for these antigens. In this study, the C4(V3)6 gene construct was introduced into lettuce using Agrobacterium-mediated transformation, and several putative transgenic lines were produced. Among those, six lines were selected and grown into the T1 generation. Levels of the expected and corrected size of the C4(V3)6 protein reached up to 240 μg/g DW in some lines.
It is important to point out that expression of a number of HIV proteins have been reported in plants including Gag, early proteins, such as Tat and Nef, as well as functional Env components (reviewed by Rosales-Mendoza et al. 2012). For example, Matoba et al. (2004) have described the design of the CTB-MPR(649-684) fusion protein, comprised of CTB and the membrane proximal (ectodomain) region of gp41 (MPR). This was transiently expressed in Nicotiana benthamiana plants. The immunogenic properties of the plant-derived CTB-MPR(649-684) were the reported by the same group, describing that it is capable to elicit systemic and mucosal immune responses against MPR(649-684) in mice when a mucosal prime-systemic boost immunization scheme was performed (Matoba et al. 2009). A number of other plant-produced recombinant proteins carrying Env epitopes have proven to be immunogenic in test animals, and capable of eliciting humoral and cellular responses. However, most of these systems rely on the use of chimeric plant virus or transient expression systems, which result in high yields, but require purification steps. In addition, oral immunogenicity of these chimeric virus or proteins has not been studied in detail (Rosales-Mendoza et al. 2012).
Therefore, findings obtained in this study serve as a major step forward in efforts to produce a reliable, efficacious, and safe plant-based vaccine against HIV. This polyvalent lettuce-derived C4(V3)6 antigen seems to be a promising candidate vaccine. However, additional studies should be conducted to assess if this antigen is capable to induce mucosal immune response in mice and non-human primates, as well as its neutralization activity against different isolates of the virus. It is expected that expression levels of C4(V3)6 accumulating in these lettuce lines would be appropriate to pursue these additional studies.
In conclusion, C4(V3)6 is correctly expressed in plants as expression platforms and it remains antigenic. The antigenic lettuce-derived C4(V3)6 constitute a promising tool for developing safe, efficient, and economic oral vaccine formulations against HIV, which could induce broad anti-HIV immune responses at mucosal levels.
References
Agrawal L, Haq W, Hanson CV, Rao DN (2003) Generating neutralizing antibodies, Th1 response and MHC non restricted immunogenicity of HIV-1 env and gag peptides in liposomes and ISCOMSs with in-built adjuvanticity. J Immune Based Ther Vaccines 1:1–5
Bradford MM (1976) A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein–dye binding. Anal Biochem 72:248–254
Bradney C, Sempowski G, Liao HX, Haynes B, Staats H (2002) Cytokines as adjuvants for the induction of anti-human immunodeficiency virus peptide immunoglobulin G (IgG) and IgA antibodies in serum and mucosal secretions after nasal immunization. J Virol 76:517–524
Curtis IS (2006) Lettuce (Lactuca sativa L.). Methods Mol Biol 343:449–458
Daniell H, StreatWeld SJ, WycoV K (2001) Medical molecular farming: production of antibodies, biopharmaceuticals and edible vaccines in plants. Trends Plant Sci 6:219–226
Dellaporta SL, Wood J, Hicks JB (1983) A plant DNA mini preparation: version II. Plant Mol Biol Rep 1:19–21
Fernández I, Gelding H, Benaissa-Trouw B, De-Vos N, Harmsen M, Nottet H, Golding B, Puijk W, Meloen R, Snippe H, Kraaijeveld C (1998) Induction of HIV-1 IIIb neutralizing antibodies in BALB/c mice by a chimaeric peptide consisting of a T-helper cell epitope of Semliki Forest virus and a B-cell epitope of HIV. Vaccine 16:1936–1940
Golding B, Eller N, Levy L, Beining P, Inman J, Matthews N, Scott DE, Golding H (2002) Mucosal immunity in mice immunized with HIV-1 peptide conjugated to Brucella abortus. Vaccine 20:1445–1450
Hamajima K, Bukawa H, Fukushima J, Kawamoto S, Kaneko T, Sekigawa K, Tanaka S, Tsukuda M, Okuda K (1995) A macromolecular multicomponent peptide vaccine prepared using the glutaraldehide conjugation method with strong immunogenicity for HIV-1. Clin Immunol Immunopathol 77:374–379
Hart MK, Plaker TJ, Matthews TJ, Langlois AJ, Lerche NW, Martin ME, Scearce RM, McDanal C, Bolognesi DP, Haynes BF (1990) Synthetic peptides containing T and B cell epitopes from human immunodeficiency virus envelope gp120 induce anti-HIV proliferative response and high titers of neutralizing antibodies in rhesus monkeys. J Immunol 145:2677–2785
Hart MK, Weinhold KJ, Scearce RM, Washburn EM, Clark CA, Palker TJ, Haynes BF (1991) Priming of anti-human immunodeficiency virus (HIV) CD8+ cytotoxic T cell in vitro by carrier-free HIV synthetic peptides. Proc Natl Acad Sci USA 88:9448–9452
Haynes B, Torres J, Langlois A, Bolognesi D, Gardner M, Palker T, Scearce R, Jones D, Moody M, McDanal C, Matthews T (1993) Induction of HIVMN neutralizing antibodies in primates using a prime-boost regimen of hybrid synthetic gp120 envelope peptides. J Immunol 151:1646–1653
Haynes BF, Benjiang M, Montefiori DC, Wrin T, Petropoulos CJ, Sutherland LL, Scearce RM, Denton C, Xia SM, Kjober BT, Liao HX (2005) Analysis of HIV-1 subtype B third variable region peptide motifs for induction of neutralizing antibodies against HIV-1 primary isolates. Virology 345:44–55
Horsch RB, Fraley RT, Rogers SG, Sanders PR, Lloyd A (1985) A simple and general method for transferring genes into plants. Science 227:1229–1231
Kanagaraj AP, Verma D, Daniell H (2011) Expression of dengue-3 premembrane and envelope polyprotein in lettuce chloroplasts. Plant Mol Biol 76:323–333
Koff WC (2012) HIV vaccine development: challenges and opportunities towards solving the HIV vaccine-neutralizing antibody problem. Vaccine 30:4310–4315
Lu Y, Xiao Y, Ding J, Dierich M, Chen YH (2000) Multiepitope vaccines intensively increased levels of antibodies recognizing three neutralizing epitopes on human immunodeficiency virus-1 envelope protein. J Immunol 51:497–501
Malm M, Rollman E, Ustay M, Hinkula J, Krohn K, Wahren B, Blazeyic V (2005) Cross-clade protection induced by human immunodeficiency virus-1 DNA immunogens expressing consensus sequences of multiple genes and epitopes from subtypes A, B, C, and FGH. Viral Immunol 18:678–688
Marcondes J, Hansen E (2008) Transgenic lettuce seedlings carrying hepatitis B virus antigen HBsAg. Braz J Infect Dis 12:469–471
Matoba N, Magérus A, Geyer BC, Zhang Y, Muralidharan M, Alfsen A, Arntzen CJ, Bomsel M, Mor TS (2004) A mucosally targeted subunit vaccine candidate eliciting HIV-1 transcytosis-blocking Abs. Proc Natl Acad Sci USA 101:13584–13589
Matoba N, Kajiura H, Cherni I, Doran JD, Bomsel M, Fujiyama K, Mor TS (2009) Biochemical and immunological characterization of the plant-derived candidate human immunodeficiency virus type 1 mucosal vaccine CTB-MPR. Plant Biotechnol J 7:129–145
Matsui T, Takita E, Sato T, Aizawa M, Ki M, Kadoyama Y, Hirano K, Kinjo S, Asao H, Kawamoto K, Kariya H, Makino S, Hamabata T, Sawada K, Kato K (2011) Production of double repeated B subunit of Shiga toxin 2e at high levels in transgenic lettuce plants as vaccine material for porcine edema disease. Transgenic Res 20:735–748
Nabel G, Makgoba W, Esparza J (2002) HIV-1 diversity and vaccine development. Science 296:2335–2339
Pniewski T, Kapusta J, Bociąg P, Wojciechowicz J, Kostrzak A, Gdula M, Fedorowicz-Strońska O, Wójcik P, Otta H, Samardakiewicz S, Wolko B, Płucienniczak A (2011) Low-dose oral immunization with lyophilized tissue of herbicide-resistant lettuce expressing hepatitis B surface antigen for prototype plant-derived vaccine tablet formulation. J Appl Genet 52:125–136
Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, Premsri N, Namwat C, de Souza M, Adams E, Benenson M, Gurunathan S, Tartaglia J, McNeil JG, Francis DP, Stablein D, Birx DL, Chunsuttiwat S, Khamboonruang C, Thongcharoen P, Robb ML, Michael NL, Kunasol P, Kim JH, MOPH-TAVEG Investigators (2009) Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med 361:2209–2220
Rosales-Mendoza S, Rubio-Infante N, Govea-Alonso D, Moreno-Fierros L (2012) Current status and perspectives of plant-based candidate vaccines against the human immunodeficiency virus (HIV). Plant Cell Rep 31:495–511
Rubio-Infante N, Govea-Alonso DO, Alpuche-Solís AG, García-Hernández AL, Soria-Guerra RE, Paz-Maldonado LMT, Ilhuicatzi-Alvarado D, Varona-Santos JT, Verdín-Terán L, Korban SS, Moreno-Fierros L, Rosales Mendoza S (2012) A chloroplast-derived C4V3 polypeptide from the human immunodeficiency virus (HIV) is orally immunogenic in mice. Plant Mol Biol 78:337–349
Smith RP, Davenport MP, Mak J (2012) The origin of genetic diversity in HIV-1. Virus Res. doi:10.1016/j.viruses.2012.06.015
Staats H, Nichols W, Palker T (1996) Mucosal immunity to HIV-1. Systemic and vaginal antibody responses after intranasal immunization with the HIV-1 C4/V3 peptide T1SP10 MN (A). J Immunol 157:462–472
Totrov M, Jiang X, Kong XP, Cohen S, Krachmarov C, Salomon A, Williams C, Seaman MS, Abagyan R, Cardozo T, Gorny MK, Wang S, Lu S, Pinter A, Zolla-Pazner S (2010) Structure-guided design and immunological characterization of immunogens presenting the HIV-1 gp123 V3 loop on a CTB scaffold. Virology 405:513–523
Twyman RM, Stoger E, Schillberg S, Christou P, Fischer R (2003) Molecular farming in plants: host systems and expression technology. Trends Biotechnol 21:570–578
Vaine M, Wang S, Liu Q, Arthos J, Montefiori D, Goepfert P, McElrath MJ, Lu S (2010) Profiles of human serum antibody responses elicited by three leading HIV vaccines focusing on the induction of Env-specific antibodies. PLoS ONE 5:1–8
Varona-Santos JT, Vázquez-Padrón R, Moreno-Fierros L (2006) Production of a short recombinant C4V3 HIV-1 Immunogen that induces strong anti-HIV responses by systemic and mucosal routes without the need of adjuvants. Viral Immunol 19:237–249
Vasan S, Michael NL (2012) Improved outlook on HIV-1 prevention and vaccine development. Expert Opin Biol Ther 12:983–994
Watkins JD, Siddappa NB, Lakhashe SK, Humbert M, Sholukh A, Hemashettar G, Wong YL, Yoon JK, Wang W, Novembre FJ, Idegbu C, Patel K, Corti D, Agatic G, Vanzetta F, Bianchi S, Heeney JL, Sallusto F, Lanzacecchia A, Ruprecht RM (2011) An anti-HIV-1 V3 loop antibody fully protects cross-clade and elicits T-cell immunity in macaques mucosally challenged with an R5 clade C SHIV. PLoS ONE 6:e18207
Zolla-Pazner S, Cohen S, Pinter A, Krachmarov C, Wrin T, Wang S, Lu S (2009) Cross-clade neutralizing antibodies against HIV-1 induced in rabbits by focusing the immune response on a neutralizing epitope. Virology 392:82–93
Zolla-Pazner S, Kong XP, Jiang X, Cardoso T, Nádas A, Cohen S, Totrov M, Seaman MS, Wang S, Lu S (2011) Cross-clade HIV-1 neutralizing antibodies induced with V3-scaffold protein immunogens following priming with gp120 DNA. J Virol 85:9887–9898
Acknowledgments
This study was funded by grants CONACYT-CB-102109, UASLP-C08-FAI-10-6.42 to SRM and PROMEP-2011 to UASLP-CA-213. Thanks are extended to Dr. Ramón García-de la Cruz for the valuable support during molecular cloning procedures and transgenic plant development.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Govea-Alonso, D.O., Gómez-Cardona, E.E., Rubio-Infante, N. et al. Production of an antigenic C4(V3)6 multiepitopic HIV protein in bacterial and plant systems. Plant Cell Tiss Organ Cult 113, 73–79 (2013). https://doi.org/10.1007/s11240-012-0252-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-012-0252-4