Abstract
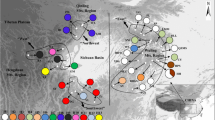
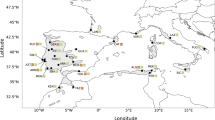
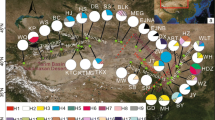
Previous phylogeographic studies of the warm-temperate zone in China focused on woody plants, but little attention was given to the climate-sensitive herbaceous plants. In this work, we implemented a phylogeographic survey on the perennial herb Achyranthes bidentata in China’s warm-temperate zone. The sequence variation of cpDNA and nDNA was examined across 209 individuals from 21 populations. A total of 11 chlorotypes and 26 ribotypes were identified. The cpDNA data showed weak population genetic differentiation and could not divide the 21 populations into different genetic groups. By contrast, the nDNA data revealed stronger genetic differentiation than cpDNA and could divide these populations into two genetic groups. The cpDNA and nDNA data both gave unambiguous signs of recent sudden population expansion. Based on the cpDNA and nDNA data, the estimated time of population expansion occurred at interglacial Marine Isotope Stage (MIS) 9 of the Penultimate Glaciation in China. The cpDNA and nDNA data suggested that the glaciation during this period deeply influenced the current distribution patterns and intraspecific divergence of A. bidentata. Our survey showed that A. bidentata tracked climatic oscillations by a large range of southward retreat into three main refugia during MIS 8, followed by the sudden northward expansion from these refugia during MIS 9.



Similar content being viewed by others
References
Avise JC (2009) Phylogeography: retrospect and prospect. J Biogeogr 36:3–15
Caicedo AL, Schaal BA (2004) Population structure and phylogeography of Solanum pimpinellifolium inferred from a nuclear gene. Mol Ecol 13:1871–1882
Chen SC, Zhang L, Zeng J, Shi F, Yang H, Mao YR, Fu CX (2012) Geographic variation of chloroplast DNA in Platycarya strobilacea (Juglandaceae). J Syst Evol 50:374–385
Chen T, Chen Q, Luo Y, Huang ZL, Zhang J, Tang HR, Pan DM, Wang XR (2015) Phylogeography of Chinese cherry (Prunus pseudocerasus Lindl.) inferred from chloroplast and nuclear DNA: insights into evolutionary patterns and demographic history. Plant Biol 17:787–797
Chiang YC, Schaal BA, Ge XJ, Chiang TY (2004) Range expansion leading to departures from neutrality in the nonsymbiotic hemoglobin gene and the cpDNA trnL-trnF intergenic spacer in Trema dielsiana (Ulmaceae). Mol Phylogenet Evol 31:929–942
Clark AG (1990) Inference of haplotypes from PCR-amplified samples of diploid populations. Mol Biol Evol 7:111–122
Clement M, Posada D, Crandall KA (2000) TCS: a computer program to estimate gene genealogies. Mol Ecol 9:1657–1660
Comes HP, Kadereit JW (1998) The effect of Quaternary climatic changes on plant distribution and evolution. Trends Plant Sci 3:432–438
Cui ZJ, Chen YX, Zhang W, Zhou SZ, Zhou LP, Zhang M, Li CC (2011) Research history, glacial chronology and origins of Quaternary glaciations in China. Quat Sci 31:749–764
Earl DA, vonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10:564–567
Fu YX (1997) Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147:915–925
Fu ZZ, Li YH, Zhang KM, Li Y (2014) Molecular data and ecological niche modeling reveal population dynamics of widespread shrub Forsythia suspensa (Oleaceae) in China’s warm-temperate zone in response to climate change during the Pleistocene. BMC Evol Biol 14:114
Hamrick JL, Godt MJW (1996) Conservation genetics of endemic plant species. In: Avise JC, Hamrick JL (eds) Conservation genetics. Chapman and Hall, New York, pp 281–304
Hamrick JL, Godt MJW, Sherman-Broyles SL (1992) Factors influencing levels of genetic diversity in woody plant species. New For 6:95–124
Harrison SP, Yu G, Takahara H, Prentice IC (2001) Palaeovegetation: diversity of temperate plants in East Asia. Nature 413:129–130
Hewitt GM (2000) The genetic legacy of the quaternary ice ages. Nature 405:907–913
Hewitt GM (2004) Genetic consequences of climatic oscillations in the quaternary. Philos Trans Roy Soc Lond B Biol Sci 359:183–195
Hickerson MJ, Carstens BC, Cavendar-Bares J, Crandall KA, Graham CH, Johnson JB, Rissler L, Victoriano PF, Yoder AD (2010) Phylogeography’s past, present and future: 10 years after Avise, 2000. Mol Phylogenet Evol 54:291–301
Jensen JL, Bohonak AJ, Kelley ST (2005) Isolation by distance, web service (version 3.16, available at: http://ibdws.sdsu.edu/). BMC Genet 6:13
Kay KM, Whittall JB, Hodges SA (2006) A survey of nuclear ribosomal internal transcribed spacer substitution rates across angiosperms: an approximate molecular clock with life history effects. BMC Evol Biol 6:36
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–134
Kress WJ, Wurdack KJ, Zimmer EA, Weigt LA, Janzen DH (2005) Use of DNA barcodes to identify flowering plants. Proc Natl Acad Sci U S A 102:8369–8374
Li JT (2008) Studies on the correlation between the struetural development of Achyranthes bidentata BL. and the accumulation of major medicinal components together with its forming of genuineness. Ph.D. Dissertation. College of Life Science, Northwest University, Xi’an
Li XH, Shao JW, Lu C, Zhang XP, Qiu YX (2012a) Chloroplast phylogeography of a temperate tree Pteroceltis tatarinowii (Ulmaceae) in China. J Syst Evol 50:325–333
Li Y, Liu P, Li YH (2012b) Intraspecific variation of Achyranthes bidentata (Amaranthaceae) in the geo-authentic product area based on internal transcribed spacer sequences of ribosomal DNA. Aust J Crop Sci 6:1655–1660
Liu JQ, Sun YS, Ge XJ, Gao LM, Qiu YX (2012) Phylogeographic studies of plants in China: advances in the past and directions in the future. J Syst Evol 50:267–275
Millien-Parra V, Jaeger JJ (1999) Island biogeography of the Japanese terrestrial mammal assemblages: an example of a relict fauna. J Biogeogr 26:959–972
Nei M, Kumar S (2000) Molecular evolution and phylogenetics. Oxford University Press, New York, pp 187–206
Nybom H (2004) Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol Ecol 13:1143–1155
Petit RJ, Duminil J, Fineschi S, Salvini D, Vendramin GG (2005) Comparative organization of chloroplast, mitochondrial and nuclear diversity in plant populations. Mol Ecol 14:689–701
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Qi XS, Chen C, Comes HP, Sakaguchi S, Liu YH, Tanaka N, Sakiom H, Qiu YX (2012) Molecular data and ecological niche modelling reveal a highly dynamic evolutionary history of the East Asian Tertiary relict Cercidiphyllum (Cercidiphyllaceae). New Phytol 196:617–630
Qiu YX, Fu CX, Comes HP (2011) Plant molecular phylogeography in China and adjacent regions: tracing the genetic imprints of Quaternary climate and environmental change in the world’s most diverse temperate flora. Mol Phylogenet Evol 59:225–244
Rogers AR, Harpending H (1992) Population growth makes waves in the distribution of pairwise genetic differences. Mol Biol Evol 9:552–569
Shafer A, Cullingham CI, CÔTÉ SD, Coltman DW (2010) Of glaciers and refugia: a decade of study sheds new light on the phylogeography of northwestern North America. Mol Ecol 19:4589–4621
Shangguan TL, Li JP, Guo DG (2009) Advance in mountain vegetation ecology in the warm-temperate zone of China. J Mt Sci 27:129–139
Shi YF, Cui ZJ, Su Z (2006) The Quaternary glaciations and environmental changes in China. Hebei Science and Technology Publishing Press, Shijiazhuang
Slatkin M (1993) Isolation by distance in equilibrium and nonequilibrium populations. Evolution 47:264–279
Smith SA, Beaulieu JM (2009) Life history influences rates of climatic niche evolution in flowering plants. Proc R Soc B 276:4345–4352
Soltis DE, Morris A, McLachlan JS, Manos PS, Soltis PS (2006) Comparative phylogeography of unglaciated eastern North America. Mol Ecol 15:4261–4293
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Wang XX, Yue JP, Sun H, Li ZM (2011) Phylogeographical study on Eriophyton wallichii (Labiatae) from alpine scree of Qinghai Tibetan Plateau. Plant Divers Resour 33:605–614
Wang W, Tian CY, Li YH, Li Y (2014) Molecular data and ecological niche modelling reveal the phylogeographic pattern of Cotinus coggygria (Anacardiaceae) in China’s warm-temperate zone. Plant Biol 16:1114–1120
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MS, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic Press, San Diego, pp 315–322
Wolfe KH, Li WH, Sharp PM (1987) Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc Natl Acad Sci U S A 84:9054–9058
Yan HF, Zhang CY, Wang FY, Hu CM, Ge XJ, Hao G (2012) Population expanding with the phalanx model and lineages split by environmental heterogeneity: a case study of Primula obconicain Subtropical China. Plos One 7:e41315
Yang L, Jiang H, Wang QH, Yang BY, Kuang HX (2012) A new feruloyl tyramine glycoside from the roots of Achyranthes bidentata. Chin J Nat Med 10:16–19
Zhang XW, Li Y, Liu CY, Xia T, Zhang Q, Fang YM (2015) Phylogeography of the temperate tree species Quercus acutissima in China: Inferences from chloroplast DNA variations. Biochem Syst Ecol 63:190–197
Zhao C, Wang CB, Ma XG, Liang QL, He XJ (2013) Phylogeographic analysis of a temperate-deciduous forest restricted plant (Bupleurum longiradiatum Turcz.) reveals two refuge areas in China with subsequent refugial isolation promoting speciation. Mol Phylogenet Evol 68:628–643
Zhu H (1997) Notes on the origin of temperate deciduous broad-leaved forests of East Asia. Bull Bot Res 17:388–396
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31100272), the Henan Agricultural University Science & Technology Innovation Fund (2016A2), the Funding Scheme of Young Backbone Teachers of Higher Education Institutions in Henan Province (2015GGJS-081), and the Key Scientific Research Projects of Henan Higher School (16A220002).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Sample: No specific permits were required for Achyranthes bidentata; all samples were collected by researchers following current Chinese regulations.
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Cai-Yun Miao and Jie Yang contributed equally to this work.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 20 kb)
Rights and permissions
About this article
Cite this article
Miao, CY., Yang, J., Mao, RL. et al. Phylogeography of Achyranthes bidentata (Amaranthaceae) in China’s Warm-Temperate Zone Inferred from Chloroplast and Nuclear DNA: Insights into Population Dynamics in Response to Climate Change During the Pleistocene. Plant Mol Biol Rep 35, 166–176 (2017). https://doi.org/10.1007/s11105-016-1013-z
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-016-1013-z