Abstract
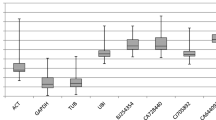
Barley (Hordeum vulgare L.) is an important cereal crop and also can be considered as a model plant for the Triticeae family. Quantitative real-time PCR (qRT-PCR) has emerged as a more practical technique to study barley gene expression, and the data normalization with reference genes is essential for accurate and reliable results. In this study, 17 candidate reference genes were evaluated from two distinct plant tissues (roots and leaves) at the barley seedling stage under different abiotic stresses (osmotic, salt, and heat) and hormonal treatments (gibberellin A3 (GA3) and methyl jasmonate (JA)). Our qRT-PCR data were analyzed with three commonly used software packages: geNorm, NormFider, and BestKeeper. In combination with the analysis of these three packages, four to five candidate genes were selected as suitable reference genes in each experimental set. These results confirmed that expression stability of reference genes depends on the experimental conditions. This study is the systematic analysis for the selection of superior reference genes for qRT-PCR in barley under different abiotic and hormonal treatments and will benefit further studies on gene expression in barley and other species of Triticeae family.



Similar content being viewed by others
References
Andersen CL, Jensen JL, Ørntoft TF (2004) Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64(15):5245–5250
Artico S, Nardeli SM, Brilhante O, Grossi-de-Sa MF, Alves-Ferreira M (2010) Identification and evaluation of new reference genes in Gossypium hirsutum for accurate normalization of real-time quantitative RT-PCR data. BMC Plant Biol 10:49. doi:10.1186/1471-2229-10-49
Brunner A, Yakovlev I, Strauss S (2004) Validating internal controls for quantitative plant gene expression studies. BMC Plant Biol 4(1):14
Bustin S (2002) Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J Mol Endocrinol 29(1):23–39
Bustin SA, Benes V, Nolan T, Pfaffl MW (2005) Quantitative real-time RT-PCR—a perspective. J Mol Endocrinol 34(3):597–601. doi:10.1677/jme.1.01755
Chen F, Wang F, Wu F, Mao W, Zhang G, Zhou M (2010) Modulation of exogenous glutathione in antioxidant defense system against Cd stress in the two barley genotypes differing in Cd tolerance. Plant Physiol Biochem 48(8):663–672
Consortium IBGS (2012) A physical, genetic and functional sequence assembly of the barley genome. Nature 491(7426):711–716
Cruz F, Kalaoun S, Nobile P, Colombo C, Almeida J, Barros LM, Romano E, Grossi-de-Sá MF, Vaslin M, Alves-Ferreira M (2009) Evaluation of coffee reference genes for relative expression studies by quantitative real-time RT-PCR. Mol Breed 23(4):607–616
Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139(1):5–17. doi:10.1104/pp. 105.063743
Derveaux S, Vandesompele J, Hellemans J (2010) How to do successful gene expression analysis using real-time PCR. Methods 50(4):227–230. doi:10.1016/j.ymeth.2009.11.001
Die JV, Roman B, Nadal S, Gonzalez-Verdejo CI (2010) Evaluation of candidate reference genes for expression studies in Pisum sativum under different experimental conditions. Planta 232(1):145–153. doi:10.1007/s00425-010-1158-1
Faccioli P, Ciceri GP, Provero P, Stanca AM, Morcia C, Terzi V (2007) A combined strategy of “in silico” transcriptome analysis and web search engine optimization allows an agile identification of reference genes suitable for normalization in gene expression studies. Plant Mol Biol 63(5):679–688. doi:10.1007/s11103-006-9116-9
Fan C, Ma J, Guo Q, Li X, Wang H, Lu M (2013) Selection of reference genes for quantitative real-time PCR in bamboo (Phyllostachys edulis). PLoS One 8(2):e56573
Goncalves S, Cairney J, Maroco J, Oliveira MM, Miguel C (2005) Evaluation of control transcripts in real-time RT-PCR expression analysis during maritime pine embryogenesis. Planta 222(3):556–563. doi:10.1007/s00425-005-1562-0
Guénin S, Mauriat M, Pelloux J, Van Wuytswinkel O, Bellini C, Gutierrez L (2009) Normalization of qRT-PCR data: the necessity of adopting a systematic, experimental conditions-specific, validation of references. J Exp Bot 60(2):487–493
Gutierrez L, Mauriat M, Guenin S, Pelloux J, Lefebvre JF, Louvet R, Rusterucci C, Moritz T, Guerineau F, Bellini C, Van Wuytswinkel O (2008a) The lack of a systematic validation of reference genes: a serious pitfall undervalued in reverse transcription-polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnol J 6(6):609–618. doi:10.1111/j.1467-7652.2008.00346.x
Gutierrez L, Mauriat M, Pelloux J, Bellini C, Van Wuytswinkel O (2008b) Towards a systematic validation of references in real-time rt-PCR. Plant Cell 20(7):1734–1735. doi:10.1105/tpc.108.059774
Hong S-Y, Seo PJ, Yang M-S, Xiang F, Park C-M (2008) Exploring valid reference genes for gene expression studies in Brachypodium distachyon by real-time PCR. BMC Plant Biol 8(1):112
Hu R, Fan C, Li H, Zhang Q, Fu YF (2009) Evaluation of putative reference genes for gene expression normalization in soybean by quantitative real-time RT-PCR. BMC Mol Biol 10:93. doi:10.1186/1471-2199-10-93
Huang J, Gu M, Lai Z, Fan B, Shi K, Zhou Y-H, Yu J-Q, Chen Z (2010) Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol 153(4):1526–1538
Huggett J, Dheda K, Bustin S, Zumla A (2005) Real-time RT-PCR normalisation; strategies and considerations. Genes Immun 6(4):279–284
Janska A, Hodek J, Svoboda P, Zamecnik J, Prasil IT, Vlasakova E, Milella L, Ovesna J (2013) The choice of reference gene set for assessing gene expression in barley (Hordeum vulgare L.) under low temperature and drought stress. Mol Genet Genomics 288(11):639–649. doi:10.1007/s00438-013-0774-4
Jarosova J, Kundu JK (2010) Validation of reference genes as internal control for studying viral infections in cereals by quantitative real-time RT-PCR. BMC Plant Biol 10:146. doi:10.1186/1471-2229-10-146
Kim B-R, Nam H-Y, Kim S-U, Kim S-I, Chang Y-J (2003) Normalization of reverse transcription quantitative-PCR with housekeeping genes in rice. Biotechnol Lett 25(21):1869–1872
Koo SC, Bracko O, Park MS, Schwab R, Chun HJ, Park KM, Seo JS, Grbic V, Balasubramanian S, Schmid M (2010) Control of lateral organ development and flowering time by the Arabidopsis thaliana MADS-box Gene AGAMOUS-LIKE6. Plant J 62(5):807–816
Kozera B, Rapacz M (2013) Reference genes in real-time PCR. J Appl Genet 54(4):391–406. doi:10.1007/s13353-013-0173-x
Kubista M, Andrade JM, Bengtsson M, Forootan A, Jonák J, Lind K, Sindelka R, Sjöback R, Sjögreen B, Strömbom L (2006) The real-time polymerase chain reaction. Mol Asp Med 27(2):95–125
Kumar V, Sharma R, Trivedi P, Vyas GK, Khandelwal V (2011) Traditional and novel references towards systematic normalization of qRT-PCR data in plants. Aust J Crop Sci 5:1455–1468
Lu J, Sivamani E, Azhakanandam K, Samadder P, Li X, Qu R (2008) Gene expression enhancement mediated by the 5′ UTR intron of the rice rubi3 gene varied remarkably among tissues in transgenic rice plants. Mol Gen Genomics 279(6):563–572
Ma S, Niu H, Liu C, Zhang J, Hou C, Wang D (2013) Expression stabilities of candidate reference genes for RT-qPCR under different stress conditions in soybean. PLoS One 8(10):e75271. doi:10.1371/journal.pone.0075271
Manoli A, Sturaro A, Trevisan S, Quaggiotti S, Nonis A (2012) Evaluation of candidate reference genes for qPCR in maize. J Plant Physiol 169(8):807–815. doi:10.1016/j.jplph.2012.01.019
Migocka M, Papierniak A (2011) Identification of suitable reference genes for studying gene expression in cucumber plants subjected to abiotic stress and growth regulators. Mol Breed 28(3):343–357
Mohammadkhani N, Heidari R (2008) Drought-induced accumulation of soluble sugars and proline in two maize varieties. World Appl Sci J 3(3):448–453
Ovesna J, Kučera L, Vaculová K, Štrymplová K, Svobodova I, Milella L (2012) Validation of the β-amy1 transcription profiling assay and selection of reference genes suited for a RT-qPCR assay in developing barley caryopsis. PLoS One 7(7):e41886
Pan L-j, Jiang L (2014) Identification and expression of the WRKY transcription factors of Carica papaya in response to abiotic and biotic stresses. Mol Biol Rep :1–11
Park S-C, Kim Y-H, Ji CY, Park S, Cheol Jeong J, Lee H-S, Kwak S-S (2012) Stable internal reference genes for the normalization of real-time PCR in different sweetpotato cultivars subjected to abiotic stress conditions. PLoS One 7(12):e51502
Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP (2004) Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnol Lett 26(6):509–515
Rapacz M, Stępień A, Skorupa K (2012) Internal standards for quantitative RT-PCR studies of gene expression under drought treatment in barley (Hordeum vulgare L.): the effects of developmental stage and leaf age. Acta Physiol Plant 34(5):1723–1733. doi:10.1007/s11738-012-0967-1
Reddy DS, Bhatnagar-Mathur P, Cindhuri KS, Sharma KK (2013) Evaluation and validation of reference genes for normalization of quantitative real-time PCR based gene expression studies in peanut. PLoS One 8(10):e78555. doi:10.1371/journal.pone.0078555
Reid KE, Olsson N, Schlosser J, Peng F, Lund ST (2006) An optimized grapevine RNA isolation procedure and statistical determination of reference genes for real-time RT-PCR during berry development. BMC Plant Biol 6(1):27
Rubie C, Kempf K, Hans J, Su T, Tilton B, Georg T, Brittner B, Ludwig B, Schilling M (2005) Housekeeping gene variability in normal and cancerous colorectal, pancreatic, esophageal, gastric and hepatic tissues. Mol Cell Probes 19(2):101–109
Schmittgen TD, Zakrajsek BA (2000) Effect of experimental treatment on housekeeping gene expression: validation by real-time, quantitative RT-PCR. J Biochem Biophys Methods 46(1):69–81
Su Y, Guo J, Ling H, Chen S, Wang S, Xu L, Allan AC, Que Y (2014) Isolation of a novel peroxisomal catalase gene from sugarcane, which is responsive to biotic and abiotic stresses. PLoS One 9(1):e84426
Tong Z, Gao Z, Wang F, Zhou J, Zhang Z (2009a) Selection of reliable reference genes for gene expression studies in peach using real-time PCR. BMC Mol Biol 10:71. doi:10.1186/1471-2199-10-71
Tong Z, Gao Z, Wang F, Zhou J, Zhang Z (2009b) Selection of reliable reference genes for gene expression studies in peach using real-time PCR. BMC Mol Biol 10(1):71
Udvardi MK, Czechowski T, Scheible W-R (2008) Eleven golden rules of quantitative RT-PCR. Plant Cell Online 20(7):1736–1737
Valasek MA, Repa JJ (2005) The power of real-time PCR. Adv Physiol Educ 29(3):151–159. doi:10.1152/advan.00019.2005
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3(7):research0034
VanGuilder HD, Vrana KE, Freeman WM (2008) Twenty-five years of quantitative PCR for gene expression analysis. Biotechniques 44(5):619
Vaucheret H, Vazquez F, Crété P, Bartel DP (2004) The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev 18(10):1187–1197
Verslues PE, Ober ES, Sharp RE (1998) Root growth and oxygen relations at low water potentials impact of oxygen availability in polyethylene glycol solutions. Plant Physiol 116:1403–1412
Walker NJ (2002) A technique whose time has come. Science 296(5567):557–559
Wan H, Zhao Z, Qian C, Sui Y, Malik AA, Chen J (2010) Selection of appropriate reference genes for gene expression studies by quantitative real-time polymerase chain reaction in cucumber. Anal Biochem 399(2):257–261. doi:10.1016/j.ab.2009.12.008
Warzybok A, Migocka M (2013) Reliable reference genes for normalization of gene expression in cucumber grown under different nitrogen nutrition. PLoS One 8(9):e72887. doi:10.1371/journal.pone.0072887
Weng X, Wang L, Wang J, Hu Y, Du H, Xu C, Xing Y, Li X, Xiao J, Zhang Q (2014) Ghd7 is a central regulator for growth, development, adaptation and responses to biotic and abiotic stresses. Plant Physiol :113.231308
Wiesner M, Hanschen FS, Schreiner M, Glatt H, Zrenner R (2013) Induced production of 1-methoxy-indol-3-ylmethyl glucosinolate by jasmonic acid and methyl jasmonate in sprouts and leaves of pak choi (Brassica rapa ssp. chinensis). Int J Mol Sci 14(7):14996–15016
Zhang L, He LL, Fu QT, Xu ZF (2013) Selection of reliable reference genes for gene expression studies in the biofuel plant Jatropha curcas using real-time quantitative PCR. Int J Mol Sci 14(12):24338–24354. doi:10.3390/ijms141224338
Zhu J, Zhang L, Li W, Han S, Yang W, Qi L (2013) Reference gene selection for quantitative real-time PCR normalization in Caragana intermedia under different abiotic stress conditions. PLoS One 8(1):e53196
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant No. 31101149), Natural Science Foundation of Zhejiang Province (Grant No. Y3110574), Public Benefit Technology Applied Research Project of Zhejiang Province (Project No. 2012C22029), China Agriculture Research System (CARS-05), the Key Research Foundation of Science and Technology Department of Zhejiang Province of China (2012C12902-2), and Agro-scientific Research in the Public Interest (201303016).
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Fig. S1
Specificity of primer pairs for qRT-PCR amplification. (A) Melting curve of the 17 candidate reference genes showing a single peak. (B) Agarose gel (2%) showing amplification of a specific PCR product of the expected size for each gene tested in this study. M: Marker 1000. (JPEG 1,532 kb)
Rights and permissions
About this article
Cite this article
Hua, W., Zhu, J., Shang, Y. et al. Identification of Suitable Reference Genes for Barley Gene Expression Under Abiotic Stresses and Hormonal Treatments. Plant Mol Biol Rep 33, 1002–1012 (2015). https://doi.org/10.1007/s11105-014-0807-0
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-014-0807-0